Differential Interaction of Antimicrobial Peptides with Lipid Structures Studied by Coarse-Grained Molecular Dynamics Simulations
- PMID: 29053635
- PMCID: PMC6151434
- DOI: 10.3390/molecules22101775
Differential Interaction of Antimicrobial Peptides with Lipid Structures Studied by Coarse-Grained Molecular Dynamics Simulations
Abstract
In this work; we investigated the differential interaction of amphiphilic antimicrobial peptides with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid structures by means of extensive molecular dynamics simulations. By using a coarse-grained (CG) model within the MARTINI force field; we simulated the peptide-lipid system from three different initial configurations: (a) peptides in water in the presence of a pre-equilibrated lipid bilayer; (b) peptides inside the hydrophobic core of the membrane; and (c) random configurations that allow self-assembled molecular structures. This last approach allowed us to sample the structural space of the systems and consider cooperative effects. The peptides used in our simulations are aurein 1.2 and maculatin 1.1; two well-known antimicrobial peptides from the Australian tree frogs; and molecules that present different membrane-perturbing behaviors. Our results showed differential behaviors for each type of peptide seen in a different organization that could guide a molecular interpretation of the experimental data. While both peptides are capable of forming membrane aggregates; the aurein 1.2 ones have a pore-like structure and exhibit a higher level of organization than those conformed by maculatin 1.1. Furthermore; maculatin 1.1 has a strong tendency to form clusters and induce curvature at low peptide-lipid ratios. The exploration of the possible lipid-peptide structures; as the one carried out here; could be a good tool for recognizing specific configurations that should be further studied with more sophisticated methodologies.
Keywords: aurein; coarse-grain; helicoidal peptides; lipid bilayers; maculatin; molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

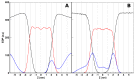

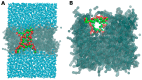

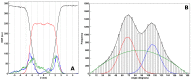


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

