The Human Knockout Gene CLYBL Connects Itaconate to Vitamin B12
- PMID: 29056341
- PMCID: PMC5827971
- DOI: 10.1016/j.cell.2017.09.051
The Human Knockout Gene CLYBL Connects Itaconate to Vitamin B12
Abstract
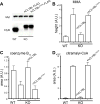
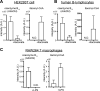
CLYBL encodes a ubiquitously expressed mitochondrial enzyme, conserved across all vertebrates, whose cellular activity and pathway assignment are unknown. Its homozygous loss is tolerated in seemingly healthy individuals, with reduced circulating B12 levels being the only and consistent phenotype reported to date. Here, by combining enzymology, structural biology, and activity-based metabolomics, we report that CLYBL operates as a citramalyl-CoA lyase in mammalian cells. Cells lacking CLYBL accumulate citramalyl-CoA, an intermediate in the C5-dicarboxylate metabolic pathway that includes itaconate, a recently identified human anti-microbial metabolite and immunomodulator. We report that CLYBL loss leads to a cell-autonomous defect in the mitochondrial B12 metabolism and that itaconyl-CoA is a cofactor-inactivating, substrate-analog inhibitor of the mitochondrial B12-dependent methylmalonyl-CoA mutase (MUT). Our work de-orphans the function of human CLYBL and reveals that a consequence of exposure to the immunomodulatory metabolite itaconate is B12 inactivation.
Keywords: C5 metabolism; CLYBL; citramalyl-CoA lyase; human genetics; itaconate; metabolism; mitochondria; systems biology; vitamin B(12).
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




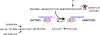
Comment in
-
A Missing Link to Vitamin B12 Metabolism.Cell. 2017 Nov 2;171(4):736-737. doi: 10.1016/j.cell.2017.10.030. Cell. 2017. PMID: 29100069
-
SnapShot: Discovering Genetic Regulatory Variants by QTL Analysis.Cell. 2017 Nov 2;171(4):980-980.e1. doi: 10.1016/j.cell.2017.10.031. Cell. 2017. PMID: 29100076 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

