Hemodynamic Forces Sculpt Developing Heart Valves through a KLF2-WNT9B Paracrine Signaling Axis
- PMID: 29056552
- PMCID: PMC5760194
- DOI: 10.1016/j.devcel.2017.09.023
Hemodynamic Forces Sculpt Developing Heart Valves through a KLF2-WNT9B Paracrine Signaling Axis
Abstract
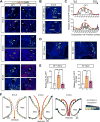
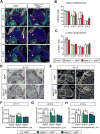
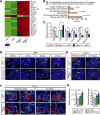
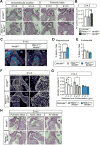
Hemodynamic forces play an essential epigenetic role in heart valve development, but how they do so is not known. Here, we show that the shear-responsive transcription factor KLF2 is required in endocardial cells to regulate the mesenchymal cell responses that remodel cardiac cushions to mature valves. Endocardial Klf2 deficiency results in defective valve formation associated with loss of Wnt9b expression and reduced canonical WNT signaling in neighboring mesenchymal cells, a phenotype reproduced by endocardial-specific loss of Wnt9b. Studies in zebrafish embryos reveal that wnt9b expression is similarly restricted to the endocardial cells overlying the developing heart valves and is dependent upon both hemodynamic shear forces and klf2a expression. These studies identify KLF2-WNT9B signaling as a conserved molecular mechanism by which fluid forces sensed by endothelial cells direct the complex cellular process of heart valve development and suggest that congenital valve defects may arise due to subtle defects in this mechanotransduction pathway.
Keywords: Klf2; Klf4; Wnt9b; cardiac cushion; endocardium; heart valve development; hemodynamic force.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



References
-
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. - PubMed
-
- Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118:649–663. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

