Transcriptome analysis reveals the genetic basis underlying the biosynthesis of volatile oil, gingerols, and diarylheptanoids in ginger (Zingiber officinale Rosc.)
- PMID: 29058093
- PMCID: PMC5651534
- DOI: 10.1186/s40529-017-0195-5
Transcriptome analysis reveals the genetic basis underlying the biosynthesis of volatile oil, gingerols, and diarylheptanoids in ginger (Zingiber officinale Rosc.)
Abstract
Background: Ginger (Zingiber officinale Rosc.) is a popular flavoring that widely used in Asian, and the volatile oil in ginger rhizomes adds a special fragrance and taste to foods. The bioactive compounds in ginger, such as gingerols, diarylheptanoids, and flavonoids, are of significant value to human health because of their anticancer, anti-oxidant, and anti-inflammatory properties. However, as a non-model plant, knowledge about the genome sequences of ginger is extremely limited, and this limits molecular studies on this plant. In this study, de novo transcriptome sequencing was performed to investigate the expression of genes associated with the biosynthesis of major bioactive compounds in matured ginger rhizome (MG), young ginger rhizome (YG), and fibrous roots of ginger (FR).
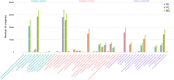
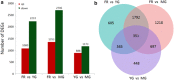
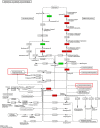
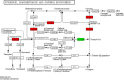
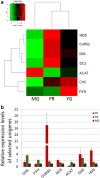
Results: A total of 361,876 unigenes were generated by de novo assembly. The expression of genes involved in the pathways responsible for the biosynthesis of major bioactive compounds differed between tissues (MG, YG, and FR). Two pathways that give rise to volatile oil, gingerols, and diarylheptanoids, the "terpenoid backbone biosynthesis" and "stilbenoid, diarylheptanoid and gingerol biosynthesis" pathways, were significantly enriched (adjusted P value < 0.05) for differentially expressed genes (DEGs) (FDR < 0.005) both between the FR and YG libraries, and the FR and MG libraries. Most of the unigenes mapped in these two pathways, including curcumin synthase, phenylpropanoylacetyl-CoA synthase, trans-cinnamate 4-monooxygenase, and 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, were expressed to a significantly higher level (log2 (fold-change) ≥ 1) in FR than in YG or MG.
Conclusion: This study provides the first insight into the biosynthesis of bioactive compounds in ginger at a molecular level and provides valuable genome resources for future molecular studies on ginger. Moreover, our results establish that bioactive compounds in ginger may predominantly synthesized in the root and then transported to rhizomes, where they accumulate.
Keywords: Bioactive compounds; Fibrous root; Ginger; Rhizome; Transcriptome sequencing.
Figures

Similar articles
-
Combined Metabolome and Transcriptome Analyses of Young, Mature, and Old Rhizome Tissues of Zingiber officinale Roscoe.Front Genet. 2021 Dec 8;12:795201. doi: 10.3389/fgene.2021.795201. eCollection 2021. Front Genet. 2021. PMID: 34956334 Free PMC article.
-
Transcriptome Analysis Provides Insights into Gingerol Biosynthesis in Ginger (Zingiber officinale).Plant Genome. 2018 Nov;11(3). doi: 10.3835/plantgenome2018.06.0034. Plant Genome. 2018. PMID: 30512040
-
Peroxisomal KAT2 (3-ketoacyl-CoA thiolase 2) gene has a key role in gingerol biosynthesis in ginger (Zingiber officinale Rosc.).J Plant Biochem Biotechnol. 2023 Jan 9:1-16. doi: 10.1007/s13562-022-00825-x. Online ahead of print. J Plant Biochem Biotechnol. 2023. PMID: 36685987 Free PMC article.
-
Ginger (Zingiber officinale Rosc.) and its bioactive components are potential resources for health beneficial agents.Phytother Res. 2021 Feb;35(2):711-742. doi: 10.1002/ptr.6858. Epub 2020 Sep 20. Phytother Res. 2021. PMID: 32954562
-
Ginger from Farmyard to Town: Nutritional and Pharmacological Applications.Front Pharmacol. 2021 Nov 26;12:779352. doi: 10.3389/fphar.2021.779352. eCollection 2021. Front Pharmacol. 2021. PMID: 34899343 Free PMC article. Review.
Cited by
-
Metabolome-driven microbiome assembly determining the health of ginger crop (Zingiber officinale L. Roscoe) against rhizome rot.Microbiome. 2024 Sep 7;12(1):167. doi: 10.1186/s40168-024-01885-y. Microbiome. 2024. PMID: 39244625 Free PMC article.
-
Soil Bacterial Community May Offer Solutions for Ginger Cultivation.Microbiol Spectr. 2022 Oct 26;10(5):e0180322. doi: 10.1128/spectrum.01803-22. Epub 2022 Sep 13. Microbiol Spectr. 2022. PMID: 36098526 Free PMC article.
-
Root diversity in sesame (Sesamum indicum L.): insights into the morphological, anatomical and gene expression profiles.Planta. 2019 Nov;250(5):1461-1474. doi: 10.1007/s00425-019-03242-y. Epub 2019 Jul 18. Planta. 2019. PMID: 31321496
-
Transcriptomic analysis of Chinese yam (Dioscorea polystachya Turcz.) variants indicates brassinosteroid involvement in tuber development.Front Nutr. 2023 May 5;10:1112793. doi: 10.3389/fnut.2023.1112793. eCollection 2023. Front Nutr. 2023. PMID: 37215221 Free PMC article.
-
Enhanced Oral Bioavailability, Anti-Tumor Activity and Hepatoprotective Effect of 6-Shogaol Loaded in a Type of Novel Micelles of Polyethylene Glycol and Linoleic Acid Conjugate.Pharmaceutics. 2019 Mar 6;11(3):107. doi: 10.3390/pharmaceutics11030107. Pharmaceutics. 2019. PMID: 30845761 Free PMC article.
References
-
- Ahmed S, Zhan CS, Yang YY, Wang XK, Yang TW, Zhao ZY, Zhang QY, Li XH, Hu XB. The transcript profile of a traditional Chinese medicine, Atractylodes lancea, revealing its sesquiterpenoid biosynthesis of the major active components. PLoS ONE. 2016;11:e0151975. doi: 10.1371/journal.pone.0151975. - DOI - PMC - PubMed
-
- Benjamini DY, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

