Preliminary comparative genomics revealed pathogenic potential and international spread of Staphylococcus argenteus
- PMID: 29058585
- PMCID: PMC5651615
- DOI: 10.1186/s12864-017-4149-9
Preliminary comparative genomics revealed pathogenic potential and international spread of Staphylococcus argenteus
Abstract
Background: Staphylococcus argenteus and S. schweitzeri, were recently proposed as novel species within S. aureus complex (SAC). S. argenteus has been reported in many countries and can threaten human health. S. schweitzeri has not been associated with human infections, but has been isolated from non-human primates. Questions regarding the evolution of pathogenicity of these two species will remain elusive until an exploratory evolutionary framework is established.
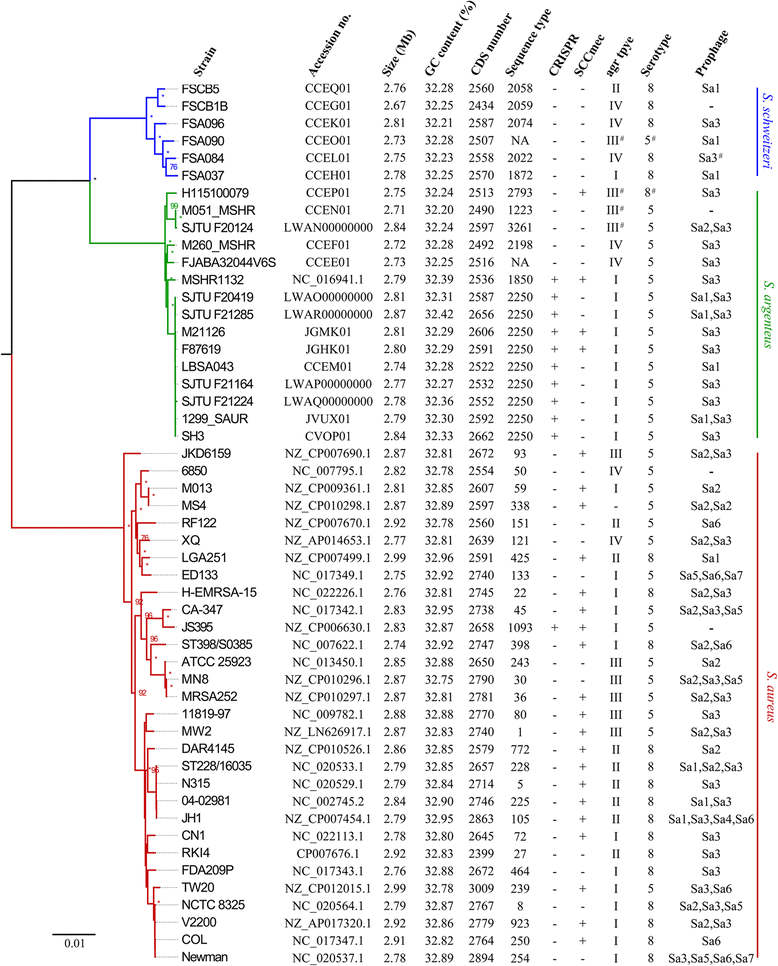
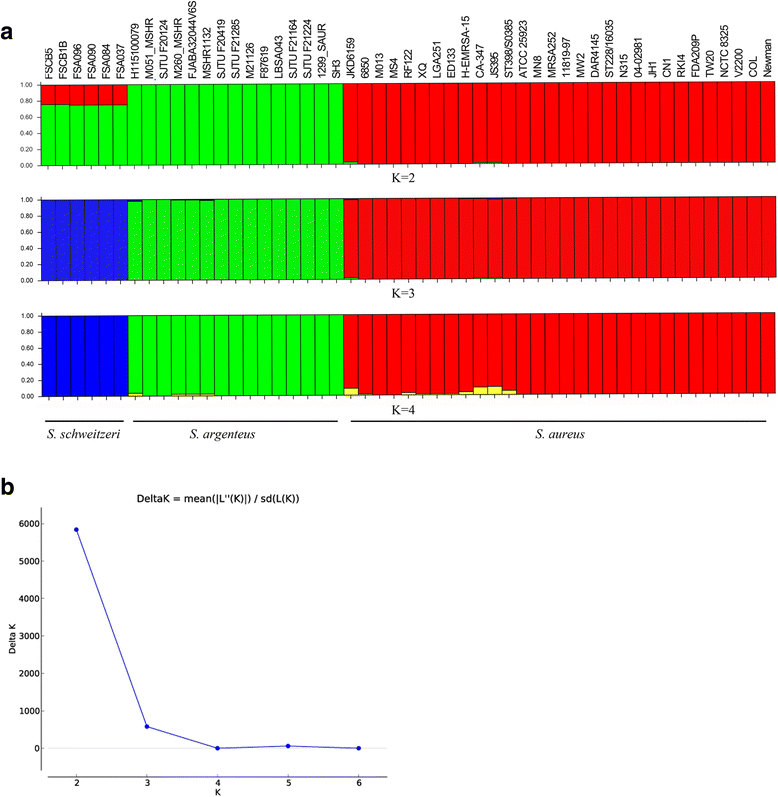
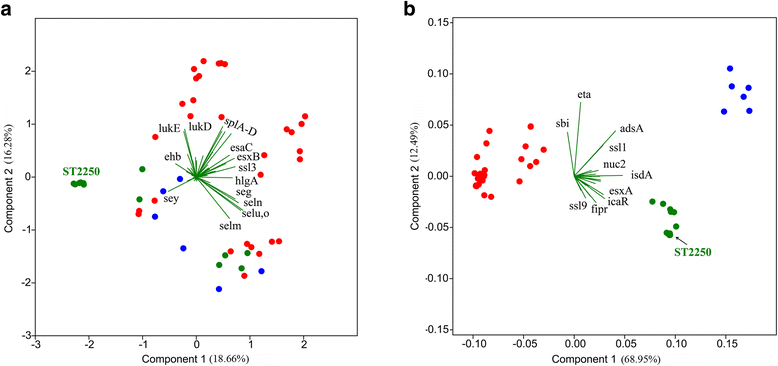
Results: We present genomic comparison analysis among members of SAC based on a pan-genome definition, which included 15 S. argenteus genomes (five newly sequenced), six S. schweitzeri genomes and 30 divergent S. aureus genomes. The three species had divergent core genomes and rare interspecific recombination was observed among the core genes. However, some subtypes of staphylococcal cassette chromosome mec (SCCmec) elements and prophages were present in different species. Of 111 tested virulence genes of S. aureus, 85 and 86 homologous genes were found in S. argenteus and S. schweitzeri, respectively. There was no difference in virulence gene content among the three species, but the sequence of most core virulence genes was divergent. Analysis of the agr locus and the genes in the capsular polysaccharides biosynthetic operon revealed that they both diverged before the speciation of SAC members. Furthermore, the widespread geographic distribution of S. argenteus, sequence type 2250, showed ambiguous biogeographical structure among geographically isolated populations, demonstrating an international spread of this pathogen.
Conclusions: S. argenteus has spread among several countries, and invasive infections and persistent carriage may be not limited to currently reported regions. S. argenteus probably had undergone a recent host adaption and can cause human infections with a similar pathogenic potential.
Keywords: Agr; Biogeographical structure; Capsular polysaccharides; Comparative genomics; Staphylococcus argenteus; Staphylococcus aureus; Staphylococcus schweitzeri; Virulence gene.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable. No ethics approval is required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Schleifer KH, Bell JA. Staphylococcus Rosenbach 1884, 18AL (nom. Cons. Opin. 17 Jud. Comm. 1958, 153.) In: De VP, editor. Bergey's manual of systematic bacteriology second edition, vol. Three, 2009. second ed. Heidelberg: Springer. p. 392–421.
-
- Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O'Brien FG, Coombs GW, Pearman JW, Tenover FC, Kapi M, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40(11):4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

