The helicase domain of Polθ counteracts RPA to promote alt-NHEJ
- PMID: 29058711
- PMCID: PMC6047744
- DOI: 10.1038/nsmb.3494
The helicase domain of Polθ counteracts RPA to promote alt-NHEJ
Abstract
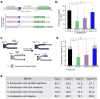
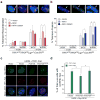
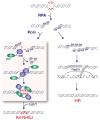
Mammalian polymerase theta (Polθ) is a multifunctional enzyme that promotes error-prone DNA repair by alternative nonhomologous end joining (alt-NHEJ). Here we present structure-function analyses that reveal that, in addition to the polymerase domain, Polθ-helicase activity plays a central role during double-strand break (DSB) repair. Our results show that the helicase domain promotes chromosomal translocations by alt-NHEJ in mouse embryonic stem cells and also suppresses CRISPR-Cas9- mediated gene targeting by homologous recombination (HR). In vitro assays demonstrate that Polθ-helicase activity facilitates the removal of RPA from resected DSBs to allow their annealing and subsequent joining by alt-NHEJ. Consistent with an antagonistic role for RPA during alt-NHEJ, inhibition of RPA1 enhances end joining and suppresses recombination. Taken together, our results reveal that the balance between HR and alt-NHEJ is controlled by opposing activities of Polθ and RPA, providing further insight into the regulation of repair-pathway choice in mammalian cells.
Conflict of interest statement
Agnel Sfeir is a founder and shareholder in Repare Therapeutics.
Figures


References
-
- Truong LN, et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

