Nutrient sensor O-GlcNAc transferase controls cancer lipid metabolism via SREBP-1 regulation
- PMID: 29059153
- PMCID: PMC5814337
- DOI: 10.1038/onc.2017.395
Nutrient sensor O-GlcNAc transferase controls cancer lipid metabolism via SREBP-1 regulation
Abstract
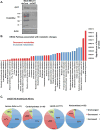
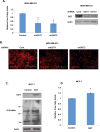
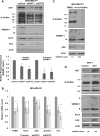
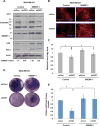
Elevated O-GlcNAcylation is associated with disease states such as diabetes and cancer. O-GlcNAc transferase (OGT) is elevated in multiple cancers and inhibition of this enzyme genetically or pharmacologically inhibits oncogenesis. Here we show that O-GlcNAcylation modulates lipid metabolism in cancer cells. OGT regulates expression of the master lipid regulator the transcription factor sterol regulatory element binding protein 1 (SREBP-1) and its transcriptional targets both in cancer and lipogenic tissue. OGT regulates SREBP-1 protein expression via AMP-activated protein kinase (AMPK). SREBP-1 is critical for OGT-mediated regulation of cell survival and of lipid synthesis, as overexpression of SREBP-1 rescues lipogenic defects associated with OGT suppression, and tumor growth in vitro and in vivo. These results unravel a previously unidentified link between O-GlcNAcylation, lipid metabolism and the regulation of SREBP-1 in cancer and suggests a crucial role for O-GlcNAc signaling in transducing nutritional state to regulate lipid metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

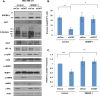
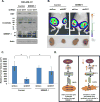
References
-
- Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007 Apr 26;446(7139):1017–22. - PubMed
-
- Varki A, Cummings R, Esko J, et al. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. The O-GlcNAc Modification.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

