The MetaCyc database of metabolic pathways and enzymes
- PMID: 29059334
- PMCID: PMC5753197
- DOI: 10.1093/nar/gkx935
The MetaCyc database of metabolic pathways and enzymes
Abstract
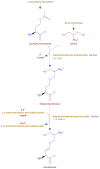
MetaCyc (https://MetaCyc.org) is a comprehensive reference database of metabolic pathways and enzymes from all domains of life. It contains more than 2570 pathways derived from >54 000 publications, making it the largest curated collection of metabolic pathways. The data in MetaCyc is strictly evidence-based and richly curated, resulting in an encyclopedic reference tool for metabolism. MetaCyc is also used as a knowledge base for generating thousands of organism-specific Pathway/Genome Databases (PGDBs), which are available in the BioCyc (https://BioCyc.org) and other PGDB collections. This article provides an update on the developments in MetaCyc during the past two years, including the expansion of data and addition of new features.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016; 44:D471–480. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

