DksA and ppGpp Regulate the σS Stress Response by Activating Promoters for the Small RNA DsrA and the Anti-Adapter Protein IraP
- PMID: 29061665
- PMCID: PMC5738727
- DOI: 10.1128/JB.00463-17
DksA and ppGpp Regulate the σS Stress Response by Activating Promoters for the Small RNA DsrA and the Anti-Adapter Protein IraP
Abstract
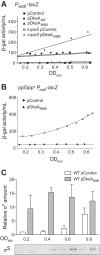
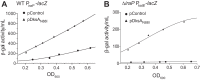
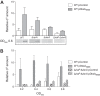
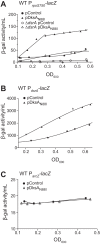
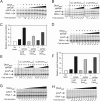
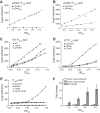
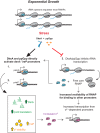
σS is an alternative sigma factor, encoded by the rpoS gene, that redirects cellular transcription to a large family of genes in response to stressful environmental signals. This so-called σS general stress response is necessary for survival in many bacterial species and is controlled by a complex, multifactorial pathway that regulates σS levels transcriptionally, translationally, and posttranslationally in Escherichia coli It was shown previously that the transcription factor DksA and its cofactor, ppGpp, are among the many factors governing σS synthesis, thus playing an important role in activation of the σS stress response. However, the mechanisms responsible for the effects of DksA and ppGpp have not been elucidated fully. We describe here how DksA and ppGpp directly activate the promoters for the anti-adaptor protein IraP and the small regulatory RNA DsrA, thereby indirectly influencing σS levels. In addition, based on effects of DksAN88I, a previously identified DksA variant with increased affinity for RNA polymerase (RNAP), we show that DksA can increase σS activity by another indirect mechanism. We propose that by reducing rRNA transcription, DksA and ppGpp increase the availability of core RNAP for binding to σS and also increase transcription from other promoters, including PdsrA and PiraP By improving the translation and stabilization of σS, as well as the ability of other promoters to compete for RNAP, DksA and ppGpp contribute to the switch in the transcription program needed for stress adaptation.IMPORTANCE Bacteria spend relatively little time in log phase outside the optimized environment found in a laboratory. They have evolved to make the most of alternating feast and famine conditions by seamlessly transitioning between rapid growth and stationary phase, a lower metabolic mode that is crucial for long-term survival. One of the key regulators of the switch in gene expression that characterizes stationary phase is the alternative sigma factor σS Understanding the factors governing σS activity is central to unraveling the complexities of growth, adaptation to stress, and pathogenesis. Here, we describe three mechanisms by which the RNA polymerase binding factor DksA and the second messenger ppGpp regulate σS levels.
Keywords: DksA; Escherichia coli; ppGpp; regulation of gene expression; small RNA; starvation; stress response.
Copyright © 2017 American Society for Microbiology.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

