IFN-stimulated Gene Expression, Type 2 Inflammation, and Endoplasmic Reticulum Stress in Asthma
- PMID: 29064281
- PMCID: PMC5811952
- DOI: 10.1164/rccm.201706-1070OC
IFN-stimulated Gene Expression, Type 2 Inflammation, and Endoplasmic Reticulum Stress in Asthma
Abstract
Rationale: Quantification of type 2 inflammation provided a molecular basis for heterogeneity in asthma. Non-type 2 pathways that contribute to asthma pathogenesis are not well understood.
Objectives: To identify dysregulated pathways beyond type 2 inflammation.
Methods: We applied RNA sequencing to airway epithelial brushings obtained from subjects with stable mild asthma not on corticosteroids (n = 19) and healthy control subjects (n = 16). Sequencing reads were mapped to human and viral genomes. In the same cohort, and in a separate group with severe asthma (n = 301), we profiled blood gene expression with microarrays.
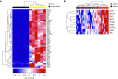
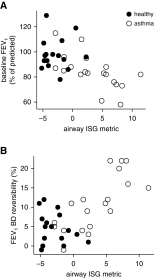
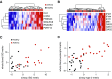
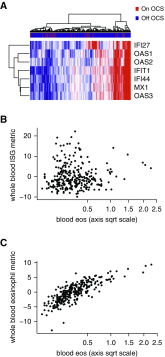
Measurements and main results: In airway brushings from mild asthma on inhaled corticosteroids, RNA sequencing yielded 1,379 differentially expressed genes (false discovery rate < 0.01). Pathway analysis revealed increased expression of type 2 markers, IFN-stimulated genes (ISGs), and endoplasmic reticulum (ER) stress-related genes. Airway epithelial ISG expression was not associated with type 2 inflammation in asthma or with viral transcripts but was associated with reduced lung function by FEV1 (ρ = -0.72; P = 0.0004). ER stress was confirmed by an increase in XBP1 (X-box binding protein 1) splicing in mild asthma and was associated with both type 2 inflammation and ISG expression. ISGs were also the most activated genes in blood cells in asthma and were correlated with airway ISG expression (ρ = 0.55; P = 0.030). High blood ISG expression in severe asthma was similarly unrelated to type 2 inflammation.
Conclusions: ISG activation is prominent in asthma, independent of viral transcripts, orthogonal to type 2 inflammation, and associated with distinct clinical features. ER stress is associated with both type 2 inflammation and ISG expression.
Keywords: IFNs; asthma; blood gene expression; endoplasmic reticulum stress; type 2 inflammation.
Figures


Comment in
-
IFN-stimulated Genes and Non-Type 2 Inflammation. New Pathway or Red Herring?Am J Respir Crit Care Med. 2018 Feb 1;197(3):277-278. doi: 10.1164/rccm.201710-2144ED. Am J Respir Crit Care Med. 2018. PMID: 29120663 Free PMC article. No abstract available.
References
-
- ’t Hoen PA, Friedländer MR, Almlöf J, Sammeth M, Pulyakhina I, Anvar SY, et al. GEUVADIS Consortium. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31:1015–1022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

