Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere
- PMID: 29064480
- PMCID: PMC5739008
- DOI: 10.1038/ismej.2017.148
Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere
Abstract
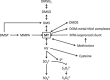
Oxidation of methanethiol (MT) is a significant step in the sulfur cycle. MT is an intermediate of metabolism of globally significant organosulfur compounds including dimethylsulfoniopropionate (DMSP) and dimethylsulfide (DMS), which have key roles in marine carbon and sulfur cycling. In aerobic bacteria, MT is degraded by a MT oxidase (MTO). The enzymatic and genetic basis of MT oxidation have remained poorly characterized. Here, we identify for the first time the MTO enzyme and its encoding gene (mtoX) in the DMS-degrading bacterium Hyphomicrobium sp. VS. We show that MTO is a homotetrameric metalloenzyme that requires Cu for enzyme activity. MTO is predicted to be a soluble periplasmic enzyme and a member of a distinct clade of the Selenium-binding protein (SBP56) family for which no function has been reported. Genes orthologous to mtoX exist in many bacteria able to degrade DMS, other one-carbon compounds or DMSP, notably in the marine model organism Ruegeria pomeroyi DSS-3, a member of the Rhodobacteraceae family that is abundant in marine environments. Marker exchange mutagenesis of mtoX disrupted the ability of R. pomeroyi to metabolize MT confirming its function in this DMSP-degrading bacterium. In R. pomeroyi, transcription of mtoX was enhanced by DMSP, methylmercaptopropionate and MT. Rates of MT degradation increased after pre-incubation of the wild-type strain with MT. The detection of mtoX orthologs in diverse bacteria, environmental samples and its abundance in a range of metagenomic data sets point to this enzyme being widely distributed in the environment and having a key role in global sulfur cycling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Antholine WE, Kastrau DHW, Steffens GCM, Buse G, Zumft WG, Kroneck PMH. (1992). A comparative EPR investigation of the multicopper proteins nitrous-oxide reductase and cytochrome-c-oxidase. Eur J Biochem 209: 875–881. - PubMed
-
- Awano S, Koshimune S, Kurihara E, Gohara K, Sakai A, Soh I et al. (2004). The assessment of methyl mercaptan, an important clinical marker for the diagnosis of oral malodor. J Dent 32: 555–559. - PubMed
-
- Bansal MP, Oborn CJ, Danielson KG, Medina D. (1989). Evidence for two selenium-binding proteins distinct from glutathione peroxidase in mouse liver. Carcinogenesis 10: 541–546. - PubMed
-
- Baumann P, Baumann L (1981). The Marine Gram Negative Eubacteria: Genera Photobacterium, Beneckea, Alteromonas, Pseudomonas and Alcaligenes. In: Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG (eds). The Prokaryotes. Springer: Berlin, pp 1302–1331..
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

