Identification of a specific peptide binding to colon cancer cells from a phage-displayed peptide library
- PMID: 29065111
- PMCID: PMC5765222
- DOI: 10.1038/bjc.2017.366
Identification of a specific peptide binding to colon cancer cells from a phage-displayed peptide library
Abstract
Background: New molecular probes are essential for early colon cancer diagnosis. A phage-display screening was performed to select novel binding peptides for early colon cancer imaging detection.
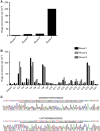
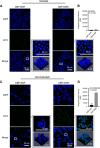
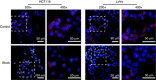
Methods: A human colon cancer cell line (COLO320HSR) and a normal human intestinal epithelial cell line (NCM460) were used for subtractive screening with a phage peptide library. The positive peptides were identified, and their binding capacities were confirmed by confocal immunofluorescence both in human colon cancer cells and in biopsy specimens. The sequences were further analysed for homology and the existing mimotopes by the BLAST algorithm and the MimoDB database.
Results: A peptide termed as CBP-DWS, which was demonstrated to be capable of binding to a panel of human colon cancer cell lines and tissues, was identified; it had virtually no binding to normal human intestinal epithelial cell line NCM460 and normal surrounding colon tissues. Bioinformatics analyses suggest that CBP-DWS targets human Glypican-3, which may be involved in important cellular functions in multiple cancer types.
Conclusions: These studies suggest that the selected peptide CBP-DWS may be a candidate to serve as a novel probe for colon cancer imaging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

