The Origin of Animal Multicellularity and Cell Differentiation
- PMID: 29065305
- PMCID: PMC6089241
- DOI: 10.1016/j.devcel.2017.09.016
The Origin of Animal Multicellularity and Cell Differentiation
Abstract
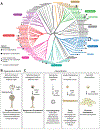
Over 600 million years ago, animals evolved from a unicellular or colonial organism whose cell(s) captured bacteria with a collar complex, a flagellum surrounded by a microvillar collar. Using principles from evolutionary cell biology, we reason that the transition to multicellularity required modification of pre-existing mechanisms for extracellular matrix synthesis and cytokinesis. We discuss two hypotheses for the origin of animal cell types: division of labor from ancient plurifunctional cells and conversion of temporally alternating phenotypes into spatially juxtaposed cell types. Mechanistic studies in diverse animals and their relatives promise to deepen our understanding of animal origins and cell biology.
Keywords: Choanozoa; choanoflagellates; evo-devo; evolutionary cell biology; metazoan origins; multicellularity.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures





References
-
- Adamska M (2016) Sponges as the rosetta stone of colonial-to-multicellular transition In: Niklas KJ, Newman SA (eds) Multicellularity. Origins and Evolution. MIT Press, pp 185–200
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

