Molecular Skin Surface-Based Transformation Visualization between Biological Macromolecules
- PMID: 29065609
- PMCID: PMC5415869
- DOI: 10.1155/2017/4818604
Molecular Skin Surface-Based Transformation Visualization between Biological Macromolecules
Abstract
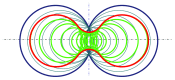
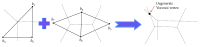
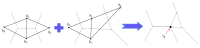
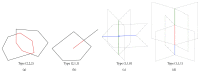
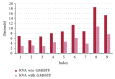
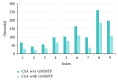
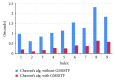
Molecular skin surface (MSS), proposed by Edelsbrunner, is a C2 continuous smooth surface modeling approach of biological macromolecules. Compared to the traditional methods of molecular surface representations (e.g., the solvent exclusive surface), MSS has distinctive advantages including having no self-intersection and being decomposable and transformable. For further promoting MSS to the field of bioinformatics, transformation between different MSS representations mimicking the macromolecular dynamics is demanded. The transformation process helps biologists understand the macromolecular dynamics processes visually in the atomic level, which is important in studying the protein structures and binding sites for optimizing drug design. However, modeling the transformation between different MSSs suffers from high computational cost while the traditional approaches reconstruct every intermediate MSS from respective intermediate union of balls. In this study, we propose a novel computational framework named general MSS transformation framework (GMSSTF) between two MSSs without the assistance of union of balls. To evaluate the effectiveness of GMSSTF, we applied it on a popular public database PDB (Protein Data Bank) and compared the existing MSS algorithms with and without GMSSTF. The simulation results show that the proposed GMSSTF effectively improves the computational efficiency and is potentially useful for macromolecular dynamic simulations.
Figures




References
-
- Bondi A. van der Waals volumes and radii. The Journal of Physical Chemistry. 1964;68(3):441–451. doi: 10.1021/j100785a001. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

