Selective Proteomic Analysis of Antibiotic-Tolerant Cellular Subpopulations in Pseudomonas aeruginosa Biofilms
- PMID: 29066549
- PMCID: PMC5654934
- DOI: 10.1128/mBio.01593-17
Selective Proteomic Analysis of Antibiotic-Tolerant Cellular Subpopulations in Pseudomonas aeruginosa Biofilms
Abstract
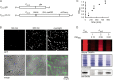
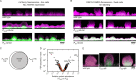
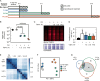
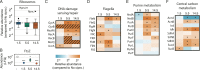
Biofilm infections exhibit high tolerance against antibiotic treatment. The study of biofilms is complicated by phenotypic heterogeneity; biofilm subpopulations differ in their metabolic activities and their responses to antibiotics. Here, we describe the use of the bio-orthogonal noncanonical amino acid tagging (BONCAT) method to enable selective proteomic analysis of a Pseudomonas aeruginosa biofilm subpopulation. Through controlled expression of a mutant methionyl-tRNA synthetase, we targeted BONCAT labeling to cells in the regions of biofilm microcolonies that showed increased tolerance to antibiotics. We enriched and identified proteins synthesized by cells in these regions. Compared to the entire biofilm proteome, the labeled subpopulation was characterized by a lower abundance of ribosomal proteins and was enriched in proteins of unknown function. We performed a pulse-labeling experiment to determine the dynamic proteomic response of the tolerant subpopulation to supra-MIC treatment with the fluoroquinolone antibiotic ciprofloxacin. The adaptive response included the upregulation of proteins required for sensing and repairing DNA damage and substantial changes in the expression of enzymes involved in central carbon metabolism. We differentiated the immediate proteomic response, characterized by an increase in flagellar motility, from the long-term adaptive strategy, which included the upregulation of purine synthesis. This targeted, selective analysis of a bacterial subpopulation demonstrates how the study of proteome dynamics can enhance our understanding of biofilm heterogeneity and antibiotic tolerance.IMPORTANCE Bacterial growth is frequently characterized by behavioral heterogeneity at the single-cell level. Heterogeneity is especially evident in the physiology of biofilms, in which distinct cellular subpopulations can respond differently to stresses, including subpopulations of pathogenic biofilms that are more tolerant to antibiotics. Global proteomic analysis affords insights into cellular physiology but cannot identify proteins expressed in a particular subpopulation of interest. Here, we report a chemical biology method to selectively label, enrich, and identify proteins expressed by cells within distinct regions of biofilm microcolonies. We used this approach to study changes in protein synthesis by the subpopulation of antibiotic-tolerant cells throughout a course of treatment. We found substantial differences between the initial response and the long-term adaptive strategy that biofilm cells use to cope with antibiotic stress. The method we describe is readily applicable to investigations of bacterial heterogeneity in diverse contexts.
Keywords: BONCAT; Pseudomonas aeruginosa; antibiotic resistance; biofilms; proteomics.
Copyright © 2017 Babin et al.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
