A novel approach for data integration and disease subtyping
- PMID: 29066617
- PMCID: PMC5741060
- DOI: 10.1101/gr.215129.116
A novel approach for data integration and disease subtyping
Abstract
Advances in high-throughput technologies allow for measurements of many types of omics data, yet the meaningful integration of several different data types remains a significant challenge. Another important and difficult problem is the discovery of molecular disease subtypes characterized by relevant clinical differences, such as survival. Here we present a novel approach, called
© 2017 Nguyen et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

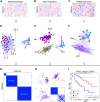


References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511. - PubMed
-
- The American Cancer Society. 2014. How is acute myeloid leukemia classified? http://www.cancer.org/cancer/leukemia-acutemyeloidaml/detailedguide/leuk....
-
- Bellman R. 1957. Dynamic programming. Princeton University Press, Princeton, NJ.
-
- Ben-Dor A, Shamir R, Yakhini Z. 1999. Clustering gene expression patterns. J Comput Biol 6: 281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
