Arabidopsis β-Amylase2 Is a K+-Requiring, Catalytic Tetramer with Sigmoidal Kinetics
- PMID: 29066669
- PMCID: PMC5717748
- DOI: 10.1104/pp.17.01506
Arabidopsis β-Amylase2 Is a K+-Requiring, Catalytic Tetramer with Sigmoidal Kinetics
Erratum in
-
CORRECTION: Vol. 175: 1525-1535, 2017.Plant Physiol. 2018 Aug;177(4):1772. doi: 10.1104/pp.18.00794. Epub 2018 Jun 28. Plant Physiol. 2018. PMID: 29954868 Free PMC article. No abstract available.
Abstract
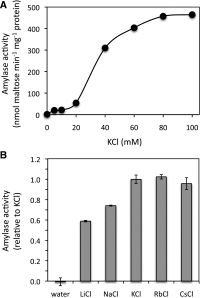
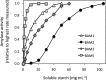
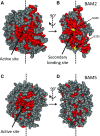
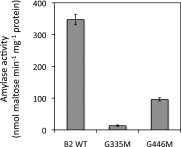
The Arabidopsis (Arabidopsis thaliana) genome contains nine β-amylase (BAM) genes, some of which play important roles in starch hydrolysis. However, little is known about BAM2, a plastid-localized enzyme reported to have extremely low catalytic activity. Using conservation of intron positions, we determined that the nine Arabidopsis BAM genes fall into two distinct subfamilies. A similar pattern was found in each major lineage of land plants, suggesting that these subfamilies diverged prior to the origin of land plants. Moreover, phylogenetic analysis indicated that BAM2 is the ancestral member of one of these subfamilies. This finding, along with the conservation of amino acids in the active site of BAM2, suggested that it might be catalytically active. We then identified KCl as necessary for BAM2 activity. Unlike BAM1, BAM3, and BAM5, three Arabidopsis BAMs that all exhibited hyperbolic kinetics, BAM2 exhibited sigmoidal kinetics with a Hill coefficient of over 3. Using multi-angle light scattering, we determined that BAM2 was a tetramer, whereas BAM5 was a monomer. Conserved residues from a diverse set of BAM2 orthologs were mapped onto a homology model of the protein, revealing a large, conserved surface away from the active site that we hypothesize is a secondary carbohydrate-binding site. Introduction of bulky methionine for glycine at two points on this surface reduced catalytic activity significantly without disrupting the tetrameric structure. Expression analysis indicated that BAM2 is more closely coexpressed with other starch degradation enzymes than any other BAM, suggesting that BAM2 may play an important role in starch degradation in plants.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures
Similar articles
-
Catalytically-inactive beta-amylase BAM4 required for starch breakdown in Arabidopsis leaves is a starch-binding-protein.Arch Biochem Biophys. 2009 Sep;489(1-2):92-8. doi: 10.1016/j.abb.2009.07.024. Epub 2009 Aug 5. Arch Biochem Biophys. 2009. PMID: 19664588
-
The BAM7 gene in Zea mays encodes a protein with similar structural and catalytic properties to Arabidopsis BAM2.Acta Crystallogr D Struct Biol. 2022 May 1;78(Pt 5):560-570. doi: 10.1107/S2059798322002169. Epub 2022 Apr 8. Acta Crystallogr D Struct Biol. 2022. PMID: 35503205 Free PMC article.
-
β-Amylase1 and β-amylase3 are plastidic starch hydrolases in Arabidopsis That Seem to Be Adapted for Different Thermal, pH, and stress conditions.Plant Physiol. 2014 Dec;166(4):1748-63. doi: 10.1104/pp.114.246421. Epub 2014 Oct 7. Plant Physiol. 2014. PMID: 25293962 Free PMC article.
-
Review: The Arabidopsis β-amylase (BAM) gene family: Diversity of form and function.Plant Sci. 2018 Nov;276:163-170. doi: 10.1016/j.plantsci.2018.08.016. Epub 2018 Aug 28. Plant Sci. 2018. PMID: 30348315 Review.
-
[New look at starch degradation in Arabidopsis thaliana L. chloroplasts].Postepy Biochem. 2007;53(1):74-83. Postepy Biochem. 2007. PMID: 17718391 Review. Polish.
Cited by
-
The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit β-amylase to promote starch degradation.Plant Cell. 2023 Dec 21;36(1):194-212. doi: 10.1093/plcell/koad259. Plant Cell. 2023. PMID: 37804098 Free PMC article.
-
Involvement of five catalytically active Arabidopsis β-amylases in leaf starch metabolism and plant growth.Plant Direct. 2020 Feb 11;4(2):e00199. doi: 10.1002/pld3.199. eCollection 2020 Feb. Plant Direct. 2020. PMID: 32072133 Free PMC article.
-
Genome-Wide Investigation of BAM Gene Family in Annona atemoya: Evolution and Expression Network Profiles during Fruit Ripening.Int J Mol Sci. 2023 Jun 22;24(13):10516. doi: 10.3390/ijms241310516. Int J Mol Sci. 2023. PMID: 37445694 Free PMC article.
-
Quaternary Structure, Salt Sensitivity, and Allosteric Regulation of β-AMYLASE2 From Arabidopsis thaliana.Front Plant Sci. 2018 Aug 14;9:1176. doi: 10.3389/fpls.2018.01176. eCollection 2018. Front Plant Sci. 2018. PMID: 30154813 Free PMC article.
-
Potassium cations expand the conformation ensemble of Arabidopsis thaliana β-amylase2 (BAM2).MicroPubl Biol. 2024 Jul 22;2024:10.17912/micropub.biology.001257. doi: 10.17912/micropub.biology.001257. eCollection 2024. MicroPubl Biol. 2024. PMID: 39109193 Free PMC article.
References
-
- Allouch J, Helbert W, Henrissat B, Czjzek M (2004) Parallel substrate binding sites in a β-agarase suggest a novel mode of action on double-helical agarose. Structure 12: 623–632 - PubMed
-
- Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry, Ed 5 WH Freeman, New York
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

