Cardamonin inhibits colonic neoplasia through modulation of MicroRNA expression
- PMID: 29066742
- PMCID: PMC5655681
- DOI: 10.1038/s41598-017-14253-8
Cardamonin inhibits colonic neoplasia through modulation of MicroRNA expression
Abstract
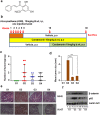
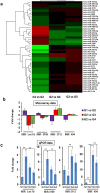
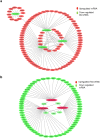
Colorectal cancer is currently the third leading cause of cancer related deaths. There is considerable interest in using dietary intervention strategies to prevent chronic diseases including cancer. Cardamonin is a spice derived nutraceutical and herein, for the first time we evaluated the therapeutic benefits of cardamonin in Azoxymethane (AOM) induced mouse model of colorectal cancer. Mice were divided into 4 groups of which three groups were given six weekly injections of AOM. One group served as untreated control and remaining groups were treated with either vehicle or Cardamonin starting from the same day or 16 weeks after the first AOM injection. Cardamonin treatment inhibited the tumor incidence, tumor multiplicity, Ki-67 and β-catenin positive cells. The activation of NF-kB signaling was also abrogated after cardamonin treatment. To elucidate the mechanism of action a global microRNA profiling of colon samples was performed. Computational analysis revealed that there is a differential expression of miRNAs between these groups. Subsequently, we extend our findings to human colorectal cancer and found that cardamonin inhibited the growth, induces cell cycle arrest and apoptosis in human colorectal cancer cell lines. Taken together, our study provides a better understanding of chemopreventive potential of cardamonin in colorectal cancer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Torre LA, et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. - PubMed
-
- Arnold, M. et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut, doi:10.1136/gutjnl-2015-310912 (2016). - PubMed
-
- Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Molecular cancer research: MCR. 2003;1:882–891. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

