DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress
- PMID: 29066820
- PMCID: PMC5654764
- DOI: 10.1038/s41467-017-01195-y
DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress
Abstract
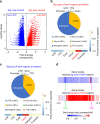
Chemical modifications on DNA molecules, such as 5-methylcytosine and 5-hydroxymethylcytosine, play important roles in the mammalian brain. A novel DNA adenine modification, N(6)-methyladenine (6mA), has recently been found in mammalian cells. However, the presence and function(s) of 6mA in the mammalian brain remain unclear. Here we demonstrate 6mA dynamics in the mouse brain in response to environmental stress. We find that overall 6mA levels are significantly elevated upon stress. Genome-wide 6mA and transcriptome profiling reveal an inverse association between 6mA dynamic changes and a set of upregulated neuronal genes or downregulated LINE transposon expression. Genes bearing stress-induced 6mA changes significantly overlap with loci associated with neuropsychiatric disorders. These results suggest an epigenetic role for 6mA in the mammalian brain as well as its potential involvement in neuropsychiatric disorders.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Similar articles
-
N6-methyladenine functions as a potential epigenetic mark in eukaryotes.Bioessays. 2015 Nov;37(11):1155-62. doi: 10.1002/bies.201500076. Epub 2015 Aug 21. Bioessays. 2015. PMID: 26293475 Review.
-
N6-methyladenine DNA modification in Drosophila.Cell. 2015 May 7;161(4):893-906. doi: 10.1016/j.cell.2015.04.018. Epub 2015 Apr 30. Cell. 2015. PMID: 25936838
-
Active N6-Methyladenine Demethylation by DMAD Regulates Gene Expression by Coordinating with Polycomb Protein in Neurons.Mol Cell. 2018 Sep 6;71(5):848-857.e6. doi: 10.1016/j.molcel.2018.07.005. Epub 2018 Aug 2. Mol Cell. 2018. PMID: 30078725 Free PMC article.
-
N6-Methyladenine DNA Methylation in Japonica and Indica Rice Genomes and Its Association with Gene Expression, Plant Development, and Stress Responses.Mol Plant. 2018 Dec 3;11(12):1492-1508. doi: 10.1016/j.molp.2018.11.005. Epub 2018 Nov 15. Mol Plant. 2018. PMID: 30448535
-
The exploration of N6-deoxyadenosine methylation in mammalian genomes.Protein Cell. 2021 Oct;12(10):756-768. doi: 10.1007/s13238-021-00866-3. Epub 2021 Aug 17. Protein Cell. 2021. PMID: 34405377 Free PMC article. Review.
Cited by
-
Juvenile stress induces behavioral change and affects perineuronal net formation in juvenile mice.BMC Neurosci. 2018 Jul 16;19(1):41. doi: 10.1186/s12868-018-0442-z. BMC Neurosci. 2018. PMID: 30012101 Free PMC article.
-
N6-Methyladenosine in RNA and DNA: An Epitranscriptomic and Epigenetic Player Implicated in Determination of Stem Cell Fate.Stem Cells Int. 2018 Oct 10;2018:3256524. doi: 10.1155/2018/3256524. eCollection 2018. Stem Cells Int. 2018. PMID: 30405719 Free PMC article. Review.
-
DNA N6-methyldeoxyadenosine in mammals and human disease.Trends Genet. 2022 May;38(5):454-467. doi: 10.1016/j.tig.2021.12.003. Epub 2022 Jan 3. Trends Genet. 2022. PMID: 34991904 Free PMC article. Review.
-
Identification of a DNA N6-Adenine Methyltransferase Complex and Its Impact on Chromatin Organization.Cell. 2019 Jun 13;177(7):1781-1796.e25. doi: 10.1016/j.cell.2019.04.028. Epub 2019 May 16. Cell. 2019. PMID: 31104845 Free PMC article.
-
Regulation of transposable elements by DNA modifications.Nat Rev Genet. 2019 Jul;20(7):417-431. doi: 10.1038/s41576-019-0106-6. Nat Rev Genet. 2019. PMID: 30867571 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

