EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies
- PMID: 29069476
- PMCID: PMC5753268
- DOI: 10.1093/nar/gkx967
EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies
Abstract
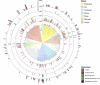
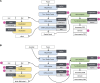
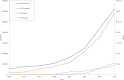
EBI metagenomics (http://www.ebi.ac.uk/metagenomics) provides a free to use platform for the analysis and archiving of sequence data derived from the microbial populations found in a particular environment. Over the past two years, EBI metagenomics has increased the number of datasets analysed 10-fold. In addition to increased throughput, the underlying analysis pipeline has been overhauled to include both new or updated tools and reference databases. Of particular note is a new workflow for taxonomic assignments that has been extended to include assignments based on both the large and small subunit RNA marker genes and to encompass all cellular micro-organisms. We also describe the addition of metagenomic assembly as a new analysis service. Our pilot studies have produced over 2400 assemblies from datasets in the public domain. From these assemblies, we have produced a searchable, non-redundant protein database of over 50 million sequences. To provide improved access to the data stored within the resource, we have developed a programmatic interface that provides access to the analysis results and associated sample metadata. Finally, we have integrated the results of a series of statistical analyses that provide estimations of diversity and sample comparisons.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Wilson M.C., Mori T., Rückert C., Uria A.R., Helf M.J., Takada K., Gernert C., Steffens U.A.E., Heycke N., Schmitt S. et al. . An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014; 506:58–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

