Predicting virus emergence amid evolutionary noise
- PMID: 29070612
- PMCID: PMC5666085
- DOI: 10.1098/rsob.170189
Predicting virus emergence amid evolutionary noise
Abstract
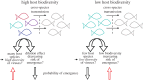
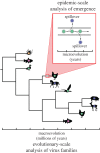
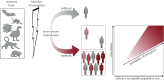
The study of virus disease emergence, whether it can be predicted and how it might be prevented, has become a major research topic in biomedicine. Here we show that efforts to predict disease emergence commonly conflate fundamentally different evolutionary and epidemiological time scales, and are likely to fail because of the enormous number of unsampled viruses that could conceivably emerge in humans. Although we know much about the patterns and processes of virus evolution on evolutionary time scales as depicted in family-scale phylogenetic trees, these data have little predictive power to reveal the short-term microevolutionary processes that underpin cross-species transmission and emergence. Truly understanding disease emergence therefore requires a new mechanistic and integrated view of the factors that allow or prevent viruses spreading in novel hosts. We present such a view, suggesting that both ecological and genetic aspects of virus emergence can be placed within a simple population genetic framework, which in turn highlights the importance of host population size and density in determining whether emergence will be successful. Despite this framework, we conclude that a more practical solution to preventing and containing the successful emergence of new diseases entails ongoing virological surveillance at the human-animal interface and regions of ecological disturbance.
Keywords: emergence; evolution; phylogeny; spill-over; virosphere; virus.
© 2017 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

