Assessment of contact predictions in CASP12: Co-evolution and deep learning coming of age
- PMID: 29071738
- PMCID: PMC5820169
- DOI: 10.1002/prot.25407
Assessment of contact predictions in CASP12: Co-evolution and deep learning coming of age
Abstract
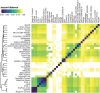
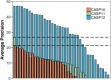
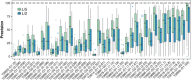
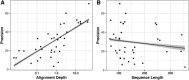
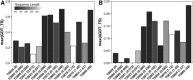
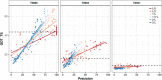
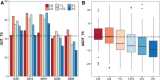
Following up on the encouraging results of residue-residue contact prediction in the CASP11 experiment, we present the analysis of predictions submitted for CASP12. The submissions include predictions of 34 groups for 38 domains classified as free modeling targets which are not accessible to homology-based modeling due to a lack of structural templates. CASP11 saw a rise of coevolution-based methods outperforming other approaches. The improvement of these methods coupled to machine learning and sequence database growth are most likely the main driver for a significant improvement in average precision from 27% in CASP11 to 47% in CASP12. In more than half of the targets, especially those with many homologous sequences accessible, precisions above 90% were achieved with the best predictors reaching a precision of 100% in some cases. We furthermore tested the impact of using these contacts as restraints in ab initio modeling of 14 single-domain free modeling targets using Rosetta. Adding contacts to the Rosetta calculations resulted in improvements of up to 26% in GDT_TS within the top five structures.
Keywords: CASP; co-variation; contact prediction; correlated mutations; de novo structure prediction; evolutionary coupling.
© 2017 The Authors Proteins: Structure, Function and Bioinformatics Published by Wiley Periodicals, Inc.
Figures

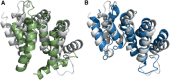


Similar articles
-
Protein contact prediction by integrating deep multiple sequence alignments, coevolution and machine learning.Proteins. 2018 Mar;86 Suppl 1(Suppl 1):84-96. doi: 10.1002/prot.25405. Epub 2017 Oct 31. Proteins. 2018. PMID: 29047157 Free PMC article.
-
Template-based and free modeling of I-TASSER and QUARK pipelines using predicted contact maps in CASP12.Proteins. 2018 Mar;86 Suppl 1(Suppl 1):136-151. doi: 10.1002/prot.25414. Epub 2017 Nov 14. Proteins. 2018. PMID: 29082551 Free PMC article.
-
Improved protein contact predictions with the MetaPSICOV2 server in CASP12.Proteins. 2018 Mar;86 Suppl 1(Suppl Suppl 1):78-83. doi: 10.1002/prot.25379. Epub 2017 Sep 29. Proteins. 2018. PMID: 28901583 Free PMC article.
-
State-of-the-art web services for de novo protein structure prediction.Brief Bioinform. 2021 May 20;22(3):bbaa139. doi: 10.1093/bib/bbaa139. Brief Bioinform. 2021. PMID: 34020540 Review.
-
Machine learning in protein structure prediction.Curr Opin Chem Biol. 2021 Dec;65:1-8. doi: 10.1016/j.cbpa.2021.04.005. Epub 2021 May 18. Curr Opin Chem Biol. 2021. PMID: 34015749 Review.
Cited by
-
Predicting the Real-Valued Inter-Residue Distances for Proteins.Adv Sci (Weinh). 2020 Aug 10;7(19):2001314. doi: 10.1002/advs.202001314. eCollection 2020 Oct. Adv Sci (Weinh). 2020. PMID: 33042750 Free PMC article.
-
Combining cysteine scanning with chemical labeling to map protein-protein interactions and infer bound structure in an intrinsically disordered region.Front Mol Biosci. 2022 Oct 7;9:997653. doi: 10.3389/fmolb.2022.997653. eCollection 2022. Front Mol Biosci. 2022. PMID: 36275627 Free PMC article.
-
Improved protein structure prediction using potentials from deep learning.Nature. 2020 Jan;577(7792):706-710. doi: 10.1038/s41586-019-1923-7. Epub 2020 Jan 15. Nature. 2020. PMID: 31942072
-
A systematic analysis of the beta hairpin motif in the Protein Data Bank.Protein Sci. 2021 Mar;30(3):613-623. doi: 10.1002/pro.4020. Epub 2021 Jan 7. Protein Sci. 2021. PMID: 33389765 Free PMC article.
-
Structure-aware protein solubility prediction from sequence through graph convolutional network and predicted contact map.J Cheminform. 2021 Feb 8;13(1):7. doi: 10.1186/s13321-021-00488-1. J Cheminform. 2021. PMID: 33557952 Free PMC article.
References
-
- Lesk AM. CASP2: report on ab initio predictions. Proteins. 1997;(suppl1):151–166. - PubMed
-
- Orengo CA, Bray JE, Hubbard T, LoConte L, Sillitoe I. Analysis and assessment of ab initio three‐dimensional prediction, secondary structure, and contacts prediction. Proteins. 1999;(suppl3):149–170. - PubMed
-
- Lesk AM, Conte Lo L, Hubbard TJ. Assessment of novel fold targets in CASP4: predictions of three‐dimensional structures, secondary structures, and interresidue contacts. Proteins. 2001;45(suppl 5):98–118. - PubMed
-
- Aloy P, Stark A, Hadley C, Russell RB. Predictions without templates: new folds, secondary structure, and contacts in CASP5. Proteins. 2003;53(suppl 6):436–456. - PubMed
-
- Graña O, Baker D, MacCallum RM, et al. CASP6 assessment of contact prediction. Proteins. 2005;61(suppl 7):214–224. - PubMed

