Novel genetic variants in the P38MAPK pathway gene ZAK and susceptibility to lung cancer
- PMID: 29071797
- PMCID: PMC6128286
- DOI: 10.1002/mc.22748
Novel genetic variants in the P38MAPK pathway gene ZAK and susceptibility to lung cancer
Abstract
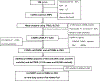
The P38MAPK pathway participates in regulating cell cycle, inflammation, development, cell death, cell differentiation, and tumorigenesis. Genetic variants of some genes in the P38MAPK pathway are reportedly associated with lung cancer risk. To substantiate this finding, we used six genome-wide association studies (GWASs) to comprehensively investigate the associations of 14 904 single nucleotide polymorphisms (SNPs) in 108 genes of this pathway with lung cancer risk. We identified six significant lung cancer risk-associated SNPs in two genes (CSNK2B and ZAK) after correction for multiple comparisons by a false discovery rate (FDR) <0.20. After removal of three CSNK2B SNPs that are located in the same locus previously reported by GWAS, we performed the LD analysis and found that rs3769201 and rs7604288 were in high LD. We then chose two independent representative SNPs of rs3769201 and rs722864 in ZAK for further analysis. We also expanded the analysis by including these two SNPs from additional GWAS datasets of Harvard University (984 cases and 970 controls) and deCODE (1319 cases and 26 380 controls). The overall effects of these two SNPs were assessed using all eight GWAS datasets (OR = 0.92, 95%CI = 0.89-0.95, and P = 1.03 × 10-5 for rs3769201; OR = 0.91, 95%CI = 0.88-0.95, and P = 2.03 × 10-6 for rs722864). Finally, we performed an expression quantitative trait loci (eQTL) analysis and found that these two SNPs were significantly associated with ZAK mRNA expression levels in lymphoblastoid cell lines. In conclusion, the ZAK rs3769201 and rs722864 may be functional susceptibility loci for lung cancer risk.
Keywords: SNP; ZAK; lung cancer risk; pathway analysis.
© 2017 Wiley Periodicals, Inc.
Figures



Similar articles
-
Genome-wide analysis of expression quantitative trait loci identified potential lung cancer susceptibility variants among Asian populations.Carcinogenesis. 2019 Apr 29;40(2):263-268. doi: 10.1093/carcin/bgy165. Carcinogenesis. 2019. PMID: 30689816
-
Potential functional variants in SMC2 and TP53 in the AURORA pathway genes and risk of pancreatic cancer.Carcinogenesis. 2019 Jun 10;40(4):521-528. doi: 10.1093/carcin/bgz029. Carcinogenesis. 2019. PMID: 30794721 Free PMC article.
-
Susceptibility loci of CNOT6 in the general mRNA degradation pathway and lung cancer risk-A re-analysis of eight GWASs.Mol Carcinog. 2017 Apr;56(4):1227-1238. doi: 10.1002/mc.22585. Epub 2016 Nov 15. Mol Carcinog. 2017. PMID: 27805284 Free PMC article.
-
Integrating expression-related SNPs into genome-wide gene- and pathway-based analyses identified novel lung cancer susceptibility genes.Int J Cancer. 2018 Apr 15;142(8):1602-1610. doi: 10.1002/ijc.31182. Epub 2017 Dec 12. Int J Cancer. 2018. PMID: 29193083
-
SNP eQTL status and eQTL density in the adjacent region of the SNP are associated with its statistical significance in GWA studies.BMC Genet. 2019 Nov 12;20(1):85. doi: 10.1186/s12863-019-0786-0. BMC Genet. 2019. PMID: 31718536 Free PMC article.
Cited by
-
Genetic variants in Hippo pathway genes are associated with house dust mite-induced allergic rhinitis in a Chinese population.Clin Transl Allergy. 2021 Dec;11(10):e12077. doi: 10.1002/clt2.12077. Clin Transl Allergy. 2021. PMID: 34962722 Free PMC article.
-
Diagnostic value of CA-153 and CYFRA 21-1 in predicting intraocular metastasis in patients with metastatic lung cancer.Cancer Med. 2020 Feb;9(4):1279-1286. doi: 10.1002/cam4.2354. Epub 2019 Jun 20. Cancer Med. 2020. PMID: 31218849 Free PMC article.
-
Gene-gene interaction of AhRwith and within the Wntcascade affects susceptibility to lung cancer.Eur J Med Res. 2022 Jan 31;27(1):14. doi: 10.1186/s40001-022-00638-7. Eur J Med Res. 2022. PMID: 35101137 Free PMC article.
-
Gallic acid suppresses the progression of triple-negative breast cancer HCC1806 cells via modulating PI3K/AKT/EGFR and MAPK signaling pathways.Front Pharmacol. 2022 Nov 29;13:1049117. doi: 10.3389/fphar.2022.1049117. eCollection 2022. Front Pharmacol. 2022. PMID: 36523491 Free PMC article.
-
Low CLOCK and CRY2 in 2nd trimester human maternal blood and risk of preterm birth: a nested case-control study†.Biol Reprod. 2021 Oct 11;105(4):827-836. doi: 10.1093/biolre/ioab119. Biol Reprod. 2021. PMID: 34142702 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA127219/CA/NCI NIH HHS/United States
- R01 CA133996/CA/NCI NIH HHS/United States
- R01 DA017932/DA/NIDA NIH HHS/United States
- U19 CA203654/CA/NCI NIH HHS/United States
- R01 CA121197/CA/NCI NIH HHS/United States
- C1298/A8780/CRUK_/Cancer Research UK/United Kingdom
- K07 CA160753/CA/NCI NIH HHS/United States
- R01 CA092039/CA/NCI NIH HHS/United States
- S10 OD018164/OD/NIH HHS/United States
- C1298/A8362/CRUK_/Cancer Research UK/United Kingdom
- U01 HG004438/HG/NHGRI NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- R01 CA111703/CA/NCI NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- P30 CA014236/CA/NCI NIH HHS/United States
- N01RC37004/RC/CCR NIH HHS/United States
- U19 CA148127/CA/NCI NIH HHS/United States
- 001/WHO_/World Health Organization/International
- R01 CA055769/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

