A global resource allocation strategy governs growth transition kinetics of Escherichia coli
- PMID: 29072300
- PMCID: PMC5901684
- DOI: 10.1038/nature24299
A global resource allocation strategy governs growth transition kinetics of Escherichia coli
Abstract
A grand challenge of systems biology is to predict the kinetic responses of living systems to perturbations starting from the underlying molecular interactions. Changes in the nutrient environment have long been used to study regulation and adaptation phenomena in microorganisms and they remain a topic of active investigation. Although much is known about the molecular interactions that govern the regulation of key metabolic processes in response to applied perturbations, they are insufficiently quantified for predictive bottom-up modelling. Here we develop a top-down approach, expanding the recently established coarse-grained proteome allocation models from steady-state growth into the kinetic regime. Using only qualitative knowledge of the underlying regulatory processes and imposing the condition of flux balance, we derive a quantitative model of bacterial growth transitions that is independent of inaccessible kinetic parameters. The resulting flux-controlled regulation model accurately predicts the time course of gene expression and biomass accumulation in response to carbon upshifts and downshifts (for example, diauxic shifts) without adjustable parameters. As predicted by the model and validated by quantitative proteomics, cells exhibit suboptimal recovery kinetics in response to nutrient shifts owing to a rigid strategy of protein synthesis allocation, which is not directed towards alleviating specific metabolic bottlenecks. Our approach does not rely on kinetic parameters, and therefore points to a theoretical framework for describing a broad range of such kinetic processes without detailed knowledge of the underlying biochemical reactions.
Conflict of interest statement
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Figures





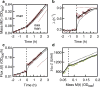






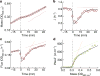
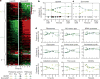
Comment in
-
Modelling the Kinetic Response to Nutrient Fluctuations.Trends Microbiol. 2018 Jan;26(1):4-5. doi: 10.1016/j.tim.2017.11.009. Epub 2017 Nov 24. Trends Microbiol. 2018. PMID: 29239769
References
-
- Monod J. The phenomenon of enzymatic adaptation and its bearings on problems of genetics and cellular differentiation. Growth. 1947;11:223–289.
-
- Kjeldgaard NO, Maaløe O, Schaechter M. The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:607–616. - PubMed
-
- Dennis PP, Bremer H. Regulation of ribonucleic acid synthesis in Escherichia coli: An analysis of a shift-up. J Mol Biol. 1973;75:145–159. - PubMed
-
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

