A bacterial toxin-antitoxin module is the origin of inter-bacterial and inter-kingdom effectors of Bartonella
- PMID: 29073136
- PMCID: PMC5675462
- DOI: 10.1371/journal.pgen.1007077
A bacterial toxin-antitoxin module is the origin of inter-bacterial and inter-kingdom effectors of Bartonella
Abstract
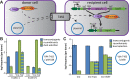
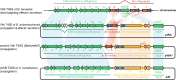
Host-targeting type IV secretion systems (T4SS) evolved from conjugative T4SS machineries that mediate interbacterial plasmid transfer. However, the origins of effectors secreted by these virulence devices have remained largely elusive. Previous work showed that some effectors exhibit homology to toxins of bacterial toxin-antitoxin modules, but the evolutionary trajectories underlying these ties had not been resolved. We previously reported that FicT toxins of FicTA toxin-antitoxin modules disrupt cellular DNA topology via their enzymatic FIC (filamentation induced by cAMP) domain. Intriguingly, the FIC domain of the FicT toxin VbhT of Bartonella schoenbuchensis is fused to a type IV secretion signal-the BID (Bep intracellular delivery) domain-similar to the Bartonella effector proteins (Beps) that are secreted into eukaryotic host cells via the host-targeting VirB T4SS. In this study, we show that the VbhT toxin is an interbacterial effector protein secreted via the conjugative Vbh T4SS that is closely related to the VirB T4SS and encoded by plasmid pVbh of B. schoenbuchensis. We therefore propose that the Vbh T4SS together with its effector VbhT represent an evolutionary missing link on a path that leads from a regular conjugation system and FicTA toxin-antitoxin modules to the VirB T4SS and the Beps. Intriguingly, phylogenetic analyses revealed that the fusion of FIC and BID domains has probably occurred independently in VbhT and the common ancestor of the Beps, suggesting parallel evolutionary paths. Moreover, several other examples of TA module toxins that are bona fide substrates of conjugative T4SS indicate that their recruitment as interbacterial effectors is prevalent and serves yet unknown biological functions in the context of bacterial conjugation. We propose that the adaptation for interbacterial transfer favors the exaptation of FicT and other TA module toxins as inter-kingdom effectors and may thus constitute an important stepping stone in the evolution of host-targeted effector proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Evolutionary Dynamics of Pathoadaptation Revealed by Three Independent Acquisitions of the VirB/D4 Type IV Secretion System in Bartonella.Genome Biol Evol. 2017 Mar 1;9(3):761-776. doi: 10.1093/gbe/evx042. Genome Biol Evol. 2017. PMID: 28338931 Free PMC article.
-
Evolutionary Diversification of Host-Targeted Bartonella Effectors Proteins Derived from a Conserved FicTA Toxin-Antitoxin Module.Microorganisms. 2021 Jul 31;9(8):1645. doi: 10.3390/microorganisms9081645. Microorganisms. 2021. PMID: 34442725 Free PMC article.
-
Parallel evolution of a type IV secretion system in radiating lineages of the host-restricted bacterial pathogen Bartonella.PLoS Genet. 2011 Feb 10;7(2):e1001296. doi: 10.1371/journal.pgen.1001296. PLoS Genet. 2011. PMID: 21347280 Free PMC article.
-
The Impact of Bartonella VirB/VirD4 Type IV Secretion System Effectors on Eukaryotic Host Cells.Front Microbiol. 2021 Dec 15;12:762582. doi: 10.3389/fmicb.2021.762582. eCollection 2021. Front Microbiol. 2021. PMID: 34975788 Free PMC article. Review.
-
Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction.Cell Microbiol. 2008 Aug;10(8):1591-8. doi: 10.1111/j.1462-5822.2008.01171.x. Epub 2008 Jul 30. Cell Microbiol. 2008. PMID: 18489724 Free PMC article. Review.
Cited by
-
Friend or Foe: Protein Inhibitors of DNA Gyrase.Biology (Basel). 2024 Jan 29;13(2):84. doi: 10.3390/biology13020084. Biology (Basel). 2024. PMID: 38392303 Free PMC article. Review.
-
Integration host factor regulates colonization factors in the bee gut symbiont Frischella perrara.Elife. 2023 Apr 14;12:e76182. doi: 10.7554/eLife.76182. Elife. 2023. PMID: 37057993 Free PMC article.
-
Bacterial injection machines: Evolutionary diverse but functionally convergent.Cell Microbiol. 2020 May;22(5):e13157. doi: 10.1111/cmi.13157. Epub 2020 Jan 16. Cell Microbiol. 2020. PMID: 31891220 Free PMC article.
-
Efficient dual-negative selection for bacterial genome editing.BMC Microbiol. 2020 May 24;20(1):129. doi: 10.1186/s12866-020-01819-2. BMC Microbiol. 2020. PMID: 32448155 Free PMC article.
-
Biology and evolution of bacterial toxin-antitoxin systems.Nat Rev Microbiol. 2022 Jun;20(6):335-350. doi: 10.1038/s41579-021-00661-1. Epub 2022 Jan 2. Nat Rev Microbiol. 2022. PMID: 34975154 Review.
References
-
- Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13(6):343–59. doi: 10.1038/nrmicro3456 . - DOI - PubMed
-
- Frank AC, Alsmark CM, Thollesson M, Andersson SG. Functional divergence and horizontal transfer of type IV secretion systems. Mol Biol Evol. 2005;22(5):1325–36. Epub 2005/03/05. doi: 10.1093/molbev/msi124 . - DOI - PubMed
-
- Abby SS, Rocha EP. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8(9):e1002983 Epub 2012/10/03. doi: 10.1371/journal.pgen.1002983 ; PubMed Central PMCID: PMCPmc3459982. - DOI - PMC - PubMed
-
- Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106(11):4154–9. doi: 10.1073/pnas.0813360106 ; PubMed Central PMCID: PMCPMC2657435. - DOI - PMC - PubMed
-
- Brown NF, Finlay BB. Potential origins and horizontal transfer of type III secretion systems and effectors. Mob Genet Elements. 2011;1(2):118–21. doi: 10.4161/mge.1.2.16733 ; PubMed Central PMCID: PMCPMC3190322. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

