BBMerge - Accurate paired shotgun read merging via overlap
- PMID: 29073143
- PMCID: PMC5657622
- DOI: 10.1371/journal.pone.0185056
BBMerge - Accurate paired shotgun read merging via overlap
Abstract
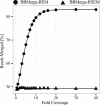
Merging paired-end shotgun reads generated on high-throughput sequencing platforms can substantially improve various subsequent bioinformatics processes, including genome assembly, binning, mapping, annotation, and clustering for taxonomic analysis. With the inexorable growth of sequence data volume and CPU core counts, the speed and scalability of read-processing tools becomes ever-more important. The accuracy of shotgun read merging is crucial as well, as errors introduced by incorrect merging percolate through to reduce the quality of downstream analysis. Thus, we designed a new tool to maximize accuracy and minimize processing time, allowing the use of read merging on larger datasets, and in analyses highly sensitive to errors. We present BBMerge, a new merging tool for paired-end shotgun sequence data. We benchmark BBMerge by comparison with eight other widely used merging tools, assessing speed, accuracy and scalability. Evaluations of both synthetic and real-world datasets demonstrate that BBMerge produces merged shotgun reads with greater accuracy and at higher speed than any existing merging tool examined. BBMerge also provides the ability to merge non-overlapping shotgun read pairs by using k-mer frequency information to assemble the unsequenced gap between reads, achieving a significantly higher merge rate while maintaining or increasing accuracy.
Conflict of interest statement
Figures





References
-
- Berka J, Chen Z, Egholm M, Godwin BC. Paired end sequencing. US Patent Office; 2009.
-
- Singer E, Andreopoulos B, Bowers RM, Lee J, Deshpande S, Chiniquy J, et al. Next generation sequencing data of a defined microbial mock community. Scientific Data. 2016;3: 160081 doi: 10.1038/sdata.2016.81 - DOI - PMC - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. Nature Publishing Group; 2001;409: 860–921. doi: 10.1038/35057062 - DOI - PubMed
-
- Ng P, Wei C-L, Sung W-K, Chiu KP, Lipovich L, Ang CC, et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat Meth. 2005;2: 105–111. doi: 10.1038/nmeth733 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

