Chemical crosslinking and mass spectrometry to elucidate the topology of integral membrane proteins
- PMID: 29073188
- PMCID: PMC5658093
- DOI: 10.1371/journal.pone.0186840
Chemical crosslinking and mass spectrometry to elucidate the topology of integral membrane proteins
Abstract
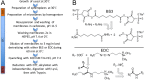
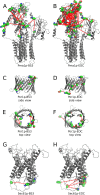
Here we made an attempt to obtain partial structural information on the topology of multispan integral membrane proteins of yeast by isolating organellar membranes, removing peripheral membrane proteins at pH 11.5 and introducing chemical crosslinks between vicinal amino acids either using homo- or hetero-bifunctional crosslinkers. Proteins were digested with specific proteases and the products analysed by mass spectrometry. Dedicated software tools were used together with filtering steps optimized to remove false positive crosslinks. In proteins of known structure, crosslinks were found only between loops residing on the same side of the membrane. As may be expected, crosslinks were mainly found in very abundant proteins. Our approach seems to hold to promise to yield low resolution topological information for naturally very abundant or strongly overexpressed proteins with relatively little effort. Here, we report novel XL-MS-based topology data for 17 integral membrane proteins (Akr1p, Fks1p, Gas1p, Ggc1p, Gpt2p, Ifa38p, Ist2p, Lag1p, Pet9p, Pma1p, Por1p, Sct1p, Sec61p, Slc1p, Spf1p, Vph1p, Ybt1p).
Conflict of interest statement
Figures



References
-
- Pagac M, de la Mora HV, Duperrex C, Roubaty C, Vionnet C, Conzelmann A. Topology of 1-acyl-sn-glycerol-3-phosphate acyltransferases SLC1 and ALE1 and related membrane-bound O-acyltransferases (MBOATs) of Saccharomyces cerevisiae. J Biol Chem. 2011;286:36438–36447. doi: 10.1074/jbc.M111.256511 - DOI - PMC - PubMed
-
- Maeda Y, Tashima Y, Houjou T, Fujita M, Yoko-o T, Jigami Y, et al. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol Biol Cell. 2007;18:1497–1506. doi: 10.1091/mbc.E06-10-0885 - DOI - PMC - PubMed
-
- Sagane K, Umemura M, Ogawa-Mitsuhashi K, Tsukahara K, Yoko-o T, Jigami Y. Analysis of membrane topology and identification of essential residues for the yeast endoplasmic reticulum inositol acyltransferase Gwt1p. J Biol Chem. 2011;286:14649–14658. doi: 10.1074/jbc.M110.193490 - DOI - PMC - PubMed
-
- Leitner A, Faini M, Stengel F, Aebersold R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem Sci. 2016;41:20–32. doi: 10.1016/j.tibs.2015.10.008 - DOI - PubMed
-
- Liu F, Heck AJ. Interrogating the architecture of protein assemblies and protein interaction networks by cross-linking mass spectrometry. Curr Opin Struct Biol. 2015;35:100–108. doi: 10.1016/j.sbi.2015.10.006 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

