Detecting the tipping points in a three-state model of complex diseases by temporal differential networks
- PMID: 29073904
- PMCID: PMC5658963
- DOI: 10.1186/s12967-017-1320-7
Detecting the tipping points in a three-state model of complex diseases by temporal differential networks
Abstract
Background: The progression of complex diseases, such as diabetes and cancer, is generally a nonlinear process with three stages, i.e., normal state, pre-disease state, and disease state, where the pre-disease state is a critical state or tipping point immediately preceding the disease state. Traditional biomarkers aim to identify a disease state by exploiting the information of differential expressions for the observed molecules, but may fail to detect a pre-disease state because there are generally little significant differences between the normal and pre-disease states. Thus, it is challenging to signal the pre-disease state, which actually implies the disease prediction.
Methods: In this work, by exploiting the information of differential associations among the observed molecules between the normal and pre-disease states, we propose a temporal differential network based computational method to accurately signal the pre-disease state or predict the occurrence of severe disease. The theoretical foundation of this work is the quantification of the critical state using dynamical network biomarkers.
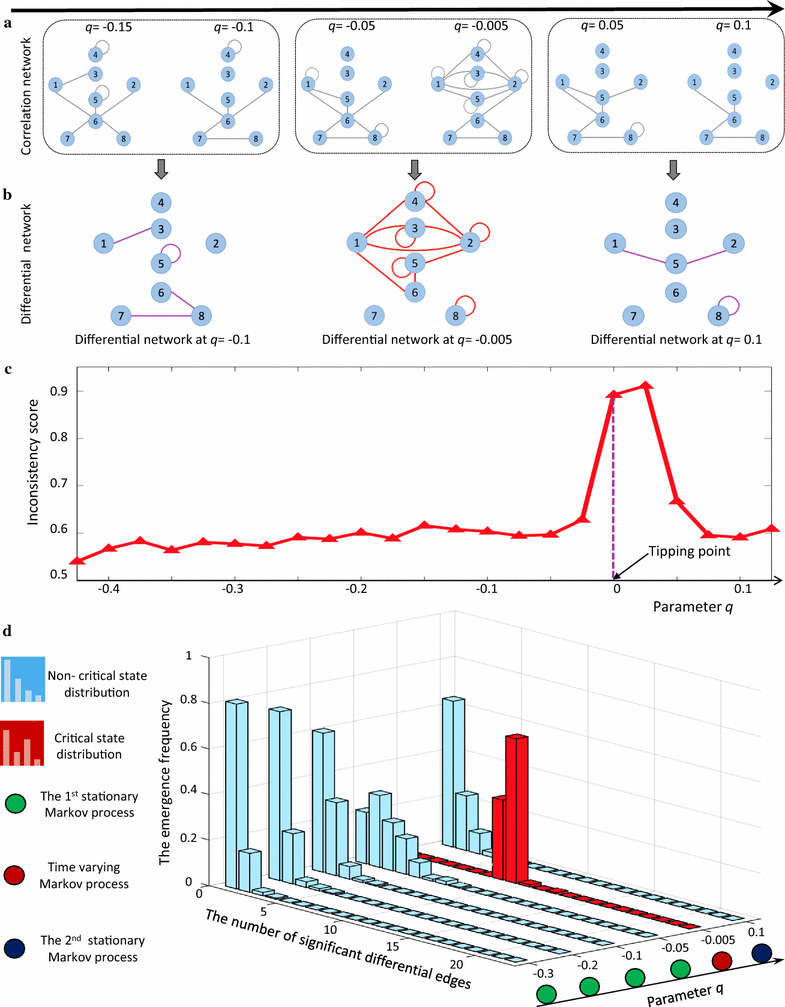
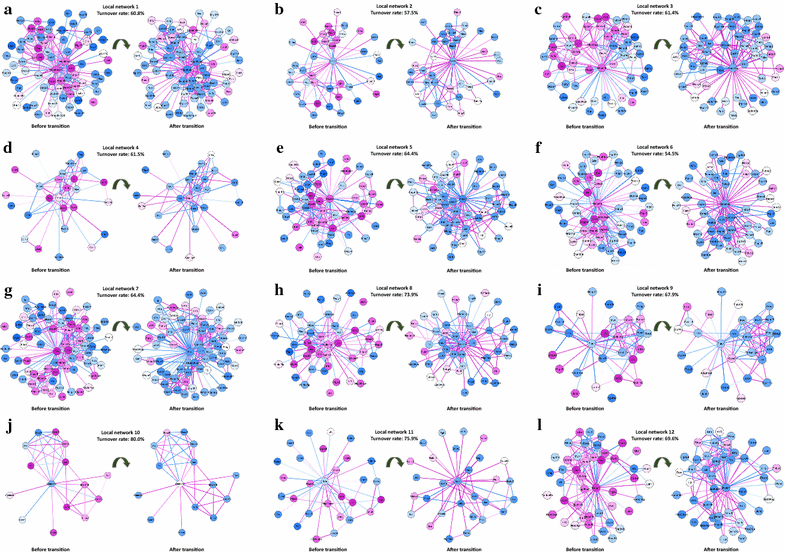
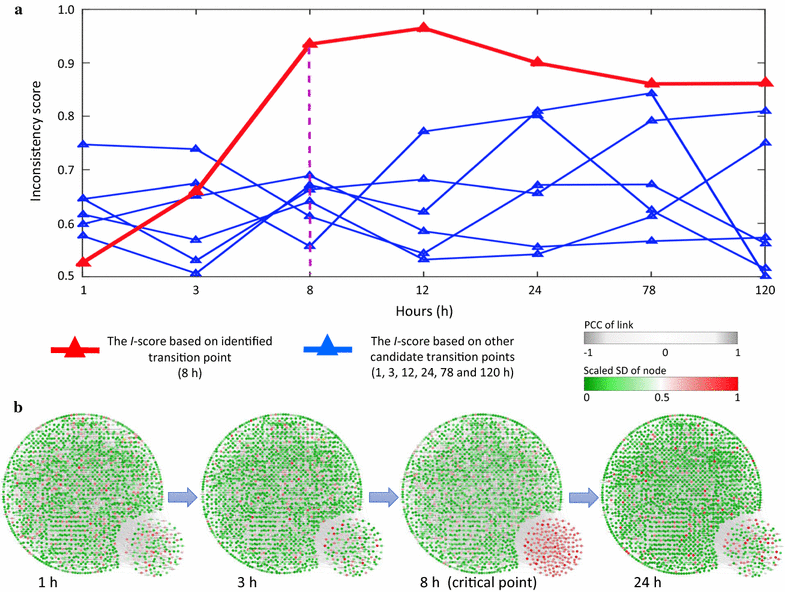
Results: Considering that there is one stationary Markov process before reaching the tipping point, a novel index, inconsistency score (I-score), is proposed to quantitatively measure the change of the stationary processes from the normal state so as to detect the onset of pre-disease state. In other words, a drastic increase of I-score implies the high inconsistency with the preceding stable state and thus signals the upcoming critical transition. This approach is applied to the simulated and real datasets of three diseases, which demonstrates the effectiveness of our method for predicting the deterioration into disease states. Both functional analysis and pathway enrichment also validate the computational results from the perspectives of both molecules and networks.
Conclusions: At the molecular network level, this method provides a computational way of unravelling the underlying mechanism of the dynamical progression when a biological system is near the tipping point, and thus detecting the early-warning signal of the imminent critical transition, which may help to achieve timely intervention. Moreover, the rewiring of differential networks effectively extracts discriminatively interpretable features, and systematically demonstrates the dynamical change of a biological system.
Keywords: Critical transition; Differential network; Dynamical network biomarker; Hidden Markov model; Pre-disease state; Tipping point.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

