S-Nitrosoglutathione Reductase Dysfunction Contributes to Obesity-Associated Hepatic Insulin Resistance via Regulating Autophagy
- PMID: 29074597
- PMCID: PMC10515702
- DOI: 10.2337/db17-0223
S-Nitrosoglutathione Reductase Dysfunction Contributes to Obesity-Associated Hepatic Insulin Resistance via Regulating Autophagy
Abstract
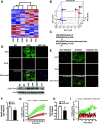
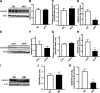
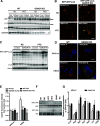
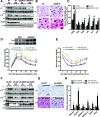
Obesity is associated with elevated intracellular nitric oxide (NO) production, which promotes nitrosative stress in metabolic tissues such as liver and skeletal muscle, contributing to insulin resistance. The onset of obesity-associated insulin resistance is due, in part, to the compromise of hepatic autophagy, a process that leads to lysosomal degradation of cellular components. However, it is not known how NO bioactivity might impact autophagy in obesity. Here, we establish that S-nitrosoglutathione reductase (GSNOR), a major protein denitrosylase, provides a key regulatory link between inflammation and autophagy, which is disrupted in obesity and diabetes. We demonstrate that obesity promotes S-nitrosylation of lysosomal proteins in the liver, thereby impairing lysosomal enzyme activities. Moreover, in mice and humans, obesity and diabetes are accompanied by decreases in GSNOR activity, engendering nitrosative stress. In mice with a GSNOR deletion, diet-induced obesity increases lysosomal nitrosative stress and impairs autophagy in the liver, leading to hepatic insulin resistance. Conversely, liver-specific overexpression of GSNOR in obese mice markedly enhances lysosomal function and autophagy and, remarkably, improves insulin action and glucose homeostasis. Furthermore, overexpression of S-nitrosylation-resistant variants of lysosomal enzymes enhances autophagy, and pharmacologically and genetically enhancing autophagy improves hepatic insulin sensitivity in GSNOR-deficient hepatocytes. Taken together, our data indicate that obesity-induced protein S-nitrosylation is a key mechanism compromising the hepatic autophagy, contributing to hepatic insulin resistance.
© 2017 by the American Diabetes Association.
Figures

Comment in
-
"NO" to Autophagy: Fat Does the Trick for Diabetes.Diabetes. 2018 Feb;67(2):180-181. doi: 10.2337/dbi17-0048. Diabetes. 2018. PMID: 29358487 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

