Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology
- PMID: 29074945
- PMCID: PMC5720686
- DOI: 10.1038/ng.3968
Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology
Abstract
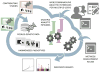
The exploding volume of whole-genome sequence (WGS) and multi-omics data requires new approaches for analysis. As one solution, we have created a cloud-based Analysis Commons, which brings together genotype and phenotype data from multiple studies in a setting that is accessible by multiple investigators. This framework addresses many of the challenges of multi-center WGS analyses, including data sharing mechanisms, phenotype harmonization, integrated multi-omics analyses, annotation, and computational flexibility. In this setting, the computational pipeline facilitates a sequence-to-discovery analysis workflow illustrated here by an analysis of plasma fibrinogen levels in 3996 individuals from the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) WGS program. The Analysis Commons represents a novel model for transforming WGS resources from a massive quantity of phenotypic and genomic data into knowledge of the determinants of health and disease risk in diverse human populations.
Figures
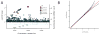
References
-
- Sankar PL, Parker LS. The Precision Medicine Initiative's All of Us Research Program: an agenda for research on its ethical, legal, and social issues. Genet Med. 2016 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- N01 HC025195/HC/NHLBI NIH HHS/United States
- R01 HL048157/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- U01 HL137181/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- U01 HL084756/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- HHSN268201500014C/HL/NHLBI NIH HHS/United States
- K23 GM102678/GM/NIGMS NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- P30 DK048520/DK/NIDDK NIH HHS/United States
- R01 HL121007/HL/NHLBI NIH HHS/United States
- P30 DK072488/DK/NIDDK NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- U54 GM115428/GM/NIGMS NIH HHS/United States
- U01 GM074518/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

