The Aromatic Head Group of Spider Toxin Polyamines Influences Toxicity to Cancer Cells
- PMID: 29077051
- PMCID: PMC5705961
- DOI: 10.3390/toxins9110346
The Aromatic Head Group of Spider Toxin Polyamines Influences Toxicity to Cancer Cells
Abstract
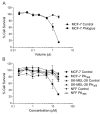
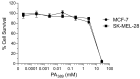
Spider venoms constitute incredibly diverse libraries of compounds, many of which are involved in prey capture and defence. Polyamines are often prevalent in the venom and target ionotropic glutamate receptors. Here we show that a novel spider polyamine, PA366, containing a hydroxyphenyl-based structure is present in the venom of several species of tarantula, and has selective toxicity against MCF-7 breast cancer cells. By contrast, a polyamine from an Australian funnel-web spider venom, which contains an identical polyamine tail to PA366 but an indole-based head-group, is only cytotoxic at high concentrations. Our results suggest that the ring structure plays a role in the cytotoxicity and that modification to the polyamine head group might lead to more potent and selective compounds with potential as novel cancer treatments.
Keywords: NMR spectroscopy; cancer; cytotoxicity; polyamine; spider venom.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Shanmugam M.K., Lee J.H., Chai E.Z., Kanchi M.M., Kar S., Arfuso F., Dharmarajan A., Kumar A.P., Ramar P.S., Looi C.Y., et al. Cancer prevention and therapy through the modulation of transcription factors by bioactive natural compounds. Semin. Cancer Biol. 2016;40–41:35–47. doi: 10.1016/j.semcancer.2016.03.005. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

