Discovering a binary CTCF code with a little help from BORIS
- PMID: 29077515
- PMCID: PMC5973138
- DOI: 10.1080/19491034.2017.1394536
Discovering a binary CTCF code with a little help from BORIS
Abstract
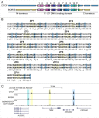
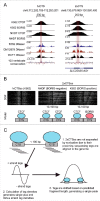
CCCTC-binding factor (CTCF) is a conserved, essential regulator of chromatin architecture containing a unique array of 11 zinc fingers (ZFs). Gene duplication and sequence divergence during early amniote evolution generated the CTCF paralog Brother Of the Regulator of Imprinted Sites (BORIS), which has a DNA binding specificity identical to that of CTCF but divergent N- and C-termini. While healthy somatic tissues express only CTCF, CTCF and BORIS are normally co-expressed in meiotic and post-meiotic germ cells, and aberrant activation of BORIS occurs in tumors and some cancer cell lines. This has led to a model in which CTCF and BORIS compete for binding to some but not all genomic target sites; however, regulation of CTCF and BORIS genomic co-occupancy is not well understood. We recently addressed this issue, finding evidence for two major classes of CTCF target sequences, some of which contain single CTCF target sites (1xCTSes) and others containing two adjacent CTCF motifs (2xCTSes). The functional and chromatin structural features of 2xCTSes are distinct from those of 1xCTS-containing regions bound by a CTCF monomer. We suggest that these previously overlooked classes of CTCF binding regions may have different roles in regulating diverse chromatin-based phenomena, and may impact our understanding of heritable epigenetic regulation in cancer cells and normal germ cells.
Keywords: 1xCTS; 2xCTS; BORIS; CTCF; CTCFL; ChIP-seq; chromatin.
Figures
References
-
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, et al.. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab‐8 insulator. EMBO Rep. 2005;6:165–70. https://doi.org/10.1038/sj.embor.7400334. PMID:15678159. - DOI - PMC - PubMed
-
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, et al.. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–24. https://doi.org/10.1128/MCB.13.12.7612. PMID:8246978. - DOI - PMC - PubMed
-
- Lutz M, Burke LJ, Barreto G, Goeman F, Greb H, Arnold R, et al.. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28:1707–13. https://doi.org/10.1093/nar/28.8.1707. PMID:10734189. - DOI - PMC - PubMed
-
- Renaud S, Loukinov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33:6850–60. https://doi.org/10.1093/nar/gki989. PMID:16326864. - DOI - PMC - PubMed
-
- Chernukhin I, Shamsuddin S, Kang SY, Bergström R, Kwon Y-W, Yu W, et al.. CTCF Interacts with and Recruits the Largest Subunit of RNA Polymerase II to CTCF Target Sites Genome-Wide. Mol Cell Biol. 2007;27:1631–48. https://doi.org/10.1128/MCB.01993-06. PMID:17210645. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
