Quantifying connectivity between local Plasmodium falciparum malaria parasite populations using identity by descent
- PMID: 29077712
- PMCID: PMC5678785
- DOI: 10.1371/journal.pgen.1007065
Quantifying connectivity between local Plasmodium falciparum malaria parasite populations using identity by descent
Abstract
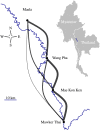
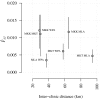
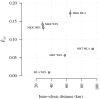
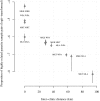
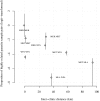
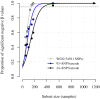
With the rapidly increasing abundance and accessibility of genomic data, there is a growing interest in using population genetic approaches to characterize fine-scale dispersal of organisms, providing insight into biological processes across a broad range of fields including ecology, evolution and epidemiology. For sexually recombining haploid organisms such as the human malaria parasite P. falciparum, however, there have been no systematic assessments of the type of data and methods required to resolve fine scale connectivity. This analytical gap hinders the use of genomics for understanding local transmission patterns, a crucial goal for policy makers charged with eliminating this important human pathogen. Here we use data collected from four clinics with a catchment area spanning approximately 120 km of the Thai-Myanmar border to compare the ability of divergence (FST) and relatedness based on identity by descent (IBD) to resolve spatial connectivity between malaria parasites collected from proximal clinics. We found no relationship between inter-clinic distance and FST, likely due to sampling of highly related parasites within clinics, but a significant decline in IBD-based relatedness with increasing inter-clinic distance. This association was contingent upon the data set type and size. We estimated that approximately 147 single-infection whole genome sequenced parasite samples or 222 single-infection parasite samples genotyped at 93 single nucleotide polymorphisms (SNPs) were sufficient to recover a robust spatial trend estimate at this scale. In summary, surveillance efforts cannot rely on classical measures of genetic divergence to measure P. falciparum transmission on a local scale. Given adequate sampling, IBD-based relatedness provides a useful alternative, and robust trends can be obtained from parasite samples genotyped at approximately 100 SNPs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Gardy J, Loman NJ, Rambaut A. Real-time digital pathogen surveillance—the time is now. Genome Biol. 2015;16(1):155 doi: 10.1186/s13059-015-0726-x - DOI - PMC - PubMed
-
- Neher RA, Bedford T. nextflu: Real-time tracking of seasonal influenza virus evolution in humans. Bioinformatics. 2015;31(21):3546–8. doi: 10.1093/bioinformatics/btv381 - DOI - PMC - PubMed
-
- Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes. Mol Biol Evol. 2016;33(8):2167–9. doi: 10.1093/molbev/msw082 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

