panX: pan-genome analysis and exploration
- PMID: 29077859
- PMCID: PMC5758898
- DOI: 10.1093/nar/gkx977
panX: pan-genome analysis and exploration
Abstract
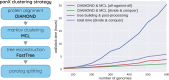
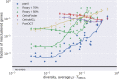
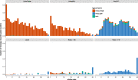
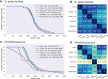
Horizontal transfer, gene loss, and duplication result in dynamic bacterial genomes shaped by a complex mixture of different modes of evolution. Closely related strains can differ in the presence or absence of many genes, and the total number of distinct genes found in a set of related isolates-the pan-genome-is often many times larger than the genome of individual isolates. We have developed a pipeline that efficiently identifies orthologous gene clusters in the pan-genome. This pipeline is coupled to a powerful yet easy-to-use web-based visualization for interactive exploration of the pan-genome. The visualization consists of connected components that allow rapid filtering and searching of genes and inspection of their evolutionary history. For each gene cluster, panX displays an alignment, a phylogenetic tree, maps mutations within that cluster to the branches of the tree and infers gain and loss of genes on the core-genome phylogeny. PanX is available at pangenome.de. Custom pan-genomes can be visualized either using a web server or by serving panX locally as a browser-based application.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Soucy S.M., Huang J., Gogarten J.P.. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015; 16:472–482. - PubMed
-
- Thomas C.M., Nielsen K.M.. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Micro. 2005; 3:711–721. - PubMed
-
- Vernikos G., Medini D., Riley D.R., Tettelin H.. Ten years of pan-genome analyses. Curr. Opin. Microbiol. 2015; 23:148–154. - PubMed
-
- Lapierre P., Gogarten J.P.. Estimating the size of the bacterial pan-genome. Trends Genet. 2009; 25:107–110. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

