Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner
- PMID: 29078276
- PMCID: PMC5676879
- DOI: 10.1073/pnas.1703921114
Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner
Abstract
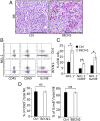
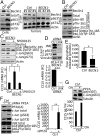
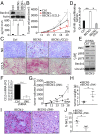
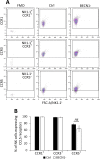
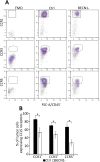
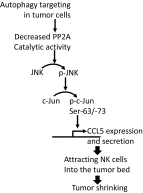
While blocking tumor growth by targeting autophagy is well established, its role on the infiltration of natural killer (NK) cells into tumors remains unknown. Here, we investigate the impact of targeting autophagy gene Beclin1 (BECN1) on the infiltration of NK cells into melanomas. We show that, in addition to inhibiting tumor growth, targeting BECN1 increased the infiltration of functional NK cells into melanoma tumors. We provide evidence that driving NK cells to the tumor bed relied on the ability of autophagy-defective tumors to transcriptionally overexpress the chemokine gene CCL5 Such infiltration and tumor regression were abrogated by silencing CCL5 in BECN1-defective tumors. Mechanistically, we show that the up-regulated expression of CCL5 occurred through the activation of its transcription factor c-Jun by a mechanism involving the impairment of phosphatase PP2A catalytic activity and the subsequent activation of JNK. Similar to BECN1, targeting other autophagy genes, such as ATG5, p62/SQSTM1, or inhibiting autophagy pharmacologically by chloroquine, also induced the expression of CCL5 in melanoma cells. Clinically, a positive correlation between CCL5 and NK cell marker NKp46 expression was found in melanoma patients, and a high expression level of CCL5 was correlated with a significant improvement of melanoma patients' survival. We believe that this study highlights the impact of targeting autophagy on the tumor infiltration by NK cells and its benefit as a novel therapeutic approach to improve NK-based immunotherapy.
Keywords: CCL5; autophagy; immunotherapy; melanoma; natural killer.
Copyright © 2017 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008;27:5932–5943. - PubMed
-
- Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17:1025–1036. - PubMed
-
- Bernardini G, Santoni A. The pathophysiological role of chemokines in the regulation of NK cell tissue homing. Crit Rev Oncog. 2014;19:77–90. - PubMed
-
- Maghazachi AA. Role of chemokines in the biology of natural killer cells. Curr Top Microbiol Immunol. 2010;341:37–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

