Trigger loop dynamics can explain stimulation of intrinsic termination by bacterial RNA polymerase without terminator hairpin contact
- PMID: 29078293
- PMCID: PMC5676890
- DOI: 10.1073/pnas.1706247114
Trigger loop dynamics can explain stimulation of intrinsic termination by bacterial RNA polymerase without terminator hairpin contact
Abstract
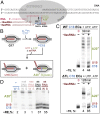
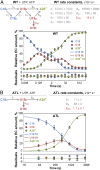
In bacteria, intrinsic termination signals cause disassembly of the highly stable elongating transcription complex (EC) over windows of two to three nucleotides after kilobases of RNA synthesis. Intrinsic termination is caused by the formation of a nascent RNA hairpin adjacent to a weak RNA-DNA hybrid within RNA polymerase (RNAP). Although the contributions of RNA and DNA sequences to termination are largely understood, the roles of conformational changes in RNAP are less well described. The polymorphous trigger loop (TL), which folds into the trigger helices to promote nucleotide addition, also is proposed to drive termination by folding into the trigger helices and contacting the terminator hairpin after invasion of the hairpin in the RNAP main cleft [Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E (2007) Mol Cell 28:991-1001]. To investigate the contribution of the TL to intrinsic termination, we developed a kinetic assay that distinguishes effects of TL alterations on the rate at which ECs terminate from effects of the TL on the nucleotide addition rate that indirectly affect termination efficiency by altering the time window in which termination can occur. We confirmed that the TL stimulates termination rate, but found that stabilizing either the folded or unfolded TL conformation decreased termination rate. We propose that conformational fluctuations of the TL (TL dynamics), not TL-hairpin contact, aid termination by increasing EC conformational diversity and thus access to favorable termination pathways. We also report that the TL and the TL sequence insertion (SI3) increase overall termination efficiency by stimulating pausing, which increases the flux of ECs into the termination pathway.
Keywords: Escherichia coli; RNA polymerase; intrinsic termination; transcription; trigger loop.
Published under the PNAS license.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Arndt KM, Chamberlin MJ. Transcription termination in Escherichia coli. Measurement of the rate of enzyme release from Rho-independent terminators. J Mol Biol. 1988;202:271–285. - PubMed
-
- Helmrich A, Ballarino M, Nudler E, Tora L. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol. 2013;20:412–418. - PubMed
-
- Wilson KS, von Hippel PH. Stability of Escherichia coli transcription complexes near an intrinsic terminator. J Mol Biol. 1994;244:36–51. - PubMed
-
- Yager TD, von Hippel PH. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991;30:1097–1118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

