Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily
- PMID: 29078300
- PMCID: PMC5692541
- DOI: 10.1073/pnas.1706849114
Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily
Abstract
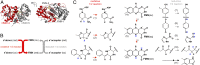
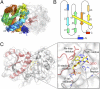
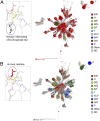
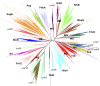
Insight regarding how diverse enzymatic functions and reactions have evolved from ancestral scaffolds is fundamental to understanding chemical and evolutionary biology, and for the exploitation of enzymes for biotechnology. We undertook an extensive computational analysis using a unique and comprehensive combination of tools that include large-scale phylogenetic reconstruction to determine the sequence, structural, and functional relationships of the functionally diverse flavin mononucleotide-dependent nitroreductase (NTR) superfamily (>24,000 sequences from all domains of life, 54 structures, and >10 enzymatic functions). Our results suggest an evolutionary model in which contemporary subgroups of the superfamily have diverged in a radial manner from a minimal flavin-binding scaffold. We identified the structural design principle for this divergence: Insertions at key positions in the minimal scaffold that, combined with the fixation of key residues, have led to functional specialization. These results will aid future efforts to delineate the emergence of functional diversity in enzyme superfamilies, provide clues for functional inference for superfamily members of unknown function, and facilitate rational redesign of the NTR scaffold.
Keywords: enzyme superfamilies; evolution; flavoenzyme; nitroreductase; sequence similarity network.
Copyright © 2017 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: Mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. - PubMed

