Complex evolutionary footprints revealed in an analysis of reused protein segments of diverse lengths
- PMID: 29078314
- PMCID: PMC5676897
- DOI: 10.1073/pnas.1707642114
Complex evolutionary footprints revealed in an analysis of reused protein segments of diverse lengths
Erratum in
-
Correction for Nepomnyachiy et al., Complex evolutionary footprints revealed in an analysis of reused protein segments of diverse lengths.Proc Natl Acad Sci U S A. 2018 Jun 5;115(23):E5430. doi: 10.1073/pnas.1807785115. Epub 2018 May 29. Proc Natl Acad Sci U S A. 2018. PMID: 29844173 Free PMC article. No abstract available.
Abstract
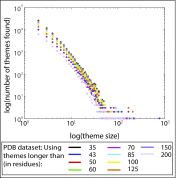
Proteins share similar segments with one another. Such "reused parts"-which have been successfully incorporated into other proteins-are likely to offer an evolutionary advantage over de novo evolved segments, as most of the latter will not even have the capacity to fold. To systematically explore the evolutionary traces of segment "reuse" across proteins, we developed an automated methodology that identifies reused segments from protein alignments. We search for "themes"-segments of at least 35 residues of similar sequence and structure-reused within representative sets of 15,016 domains [Evolutionary Classification of Protein Domains (ECOD) database] or 20,398 chains [Protein Data Bank (PDB)]. We observe that theme reuse is highly prevalent and that reuse is more extensive when the length threshold for identifying a theme is lower. Structural domains, the best characterized form of reuse in proteins, are just one of many complex and intertwined evolutionary traces. Others include long themes shared among a few proteins, which encompass and overlap with shorter themes that recur in numerous proteins. The observed complexity is consistent with evolution by duplication and divergence, and some of the themes might include descendants of ancestral segments. The observed recursive footprints, where the same amino acid can simultaneously participate in several intertwined themes, could be a useful concept for protein design. Data are available at http://trachel-srv.cs.haifa.ac.il/rachel/ppi/themes/.
Keywords: ancestral segments; protein evolutionary patterns; protein function annotation; protein space.
Copyright © 2017 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lupas AN, Ponting CP, Russell RB. On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J Struct Biol. 2001;134:191–203. - PubMed
-
- Söding J, Lupas AN. More than the sum of their parts: On the evolution of proteins from peptides. Bioessays. 2003;25:837–846. - PubMed
-
- Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14:208–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

