Algorithm for cellular reprogramming
- PMID: 29078370
- PMCID: PMC5692574
- DOI: 10.1073/pnas.1712350114
Algorithm for cellular reprogramming
Abstract
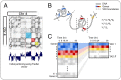
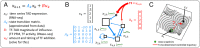
The day we understand the time evolution of subcellular events at a level of detail comparable to physical systems governed by Newton's laws of motion seems far away. Even so, quantitative approaches to cellular dynamics add to our understanding of cell biology. With data-guided frameworks we can develop better predictions about, and methods for, control over specific biological processes and system-wide cell behavior. Here we describe an approach for optimizing the use of transcription factors (TFs) in cellular reprogramming, based on a device commonly used in optimal control. We construct an approximate model for the natural evolution of a cell-cycle-synchronized population of human fibroblasts, based on data obtained by sampling the expression of 22,083 genes at several time points during the cell cycle. To arrive at a model of moderate complexity, we cluster gene expression based on division of the genome into topologically associating domains (TADs) and then model the dynamics of TAD expression levels. Based on this dynamical model and additional data, such as known TF binding sites and activity, we develop a methodology for identifying the top TF candidates for a specific cellular reprogramming task. Our data-guided methodology identifies a number of TFs previously validated for reprogramming and/or natural differentiation and predicts some potentially useful combinations of TFs. Our findings highlight the immense potential of dynamical models, mathematics, and data-guided methodologies for improving strategies for control over biological processes.
Keywords: cellular reprogramming; control theory; genome architecture; networks; time series data.
Copyright © 2017 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
-
- Brockett R. Finite Dimensional Linear Systems. Wiley; New York: 1970.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

