Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae
- PMID: 29078371
- PMCID: PMC5692575
- DOI: 10.1073/pnas.1712396114
Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae
Abstract
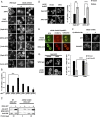
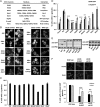
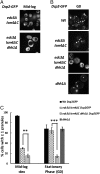
Eukaryotic cells contain multiple RNA-protein assemblies referred to as RNP granules, which are thought to form through multiple protein-protein interactions analogous to a liquid-liquid phase separation. One class of RNP granules consists of P bodies, which consist of nontranslating mRNAs and the general translation repression and mRNA degradation machinery. P bodies have been suggested to form predominantly through interactions of Edc3 and a prion-like domain on Lsm4. In this work, we provide evidence that P-body assembly can be driven by multiple different protein-protein and/or protein-RNA interactions, including interactions involving Dhh1, Psp2, and Pby1. Moreover, the relative importance of specific interactions can vary with different growth conditions. Based on these observations, we develop a summative model wherein the P-body assembly phenotype of a given mutant can be predicted from the number of currently known protein-protein interactions between P-body components.
Keywords: P bodies; RNP granules; mRNA decay.
Published under the PNAS license.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Jain S, Parker R. The discovery and analysis of P bodies. Adv Exp Med Biol. 2013;768:23–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

