WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy
- PMID: 29078390
- PMCID: PMC5676932
- DOI: 10.1073/pnas.1713625114
WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy
Abstract
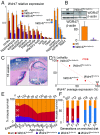
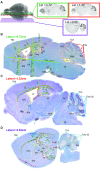
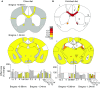
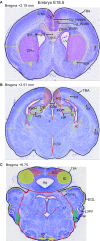
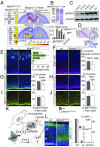
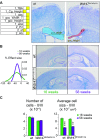
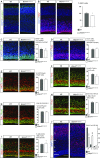
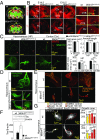
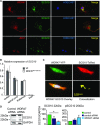
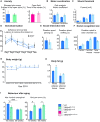
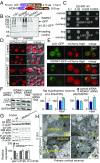
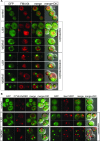
The family of WD40-repeat (WDR) proteins is one of the largest in eukaryotes, but little is known about their function in brain development. Among 26 WDR genes assessed, we found 7 displaying a major impact in neuronal morphology when inactivated in mice. Remarkably, all seven genes showed corpus callosum defects, including thicker (Atg16l1, Coro1c, Dmxl2, and Herc1), thinner (Kif21b and Wdr89), or absent corpus callosum (Wdr47), revealing a common role for WDR genes in brain connectivity. We focused on the poorly studied WDR47 protein sharing structural homology with LIS1, which causes lissencephaly. In a dosage-dependent manner, mice lacking Wdr47 showed lethality, extensive fiber defects, microcephaly, thinner cortices, and sensory motor gating abnormalities. We showed that WDR47 shares functional characteristics with LIS1 and participates in key microtubule-mediated processes, including neural stem cell proliferation, radial migration, and growth cone dynamics. In absence of WDR47, the exhaustion of late cortical progenitors and the consequent decrease of neurogenesis together with the impaired survival of late-born neurons are likely yielding to the worsening of the microcephaly phenotype postnatally. Interestingly, the WDR47-specific C-terminal to LisH (CTLH) domain was associated with functions in autophagy described in mammals. Silencing WDR47 in hypothalamic GT1-7 neuronal cells and yeast models independently recapitulated these findings, showing conserved mechanisms. Finally, our data identified superior cervical ganglion-10 (SCG10) as an interacting partner of WDR47. Taken together, these results provide a starting point for studying the implications of WDR proteins in neuronal regulation of microtubules and autophagy.
Keywords: WD40-repeat proteins; autophagy; corpus callosum agenesis; microcephaly; neurogenesis.
Published under the PNAS license.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: Structures, functions, and evolution. J Struct Biol. 2001;134:117–131. - PubMed
-
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature. 1996;379:369–374. - PubMed
-
- Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–574. - PubMed
-
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: A common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. - PubMed
-
- Hurtley S. Spatial cell biology. Location, location, location. Introduction. Science. 2009;326:1205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

