Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction
- PMID: 29078754
- PMCID: PMC5658915
- DOI: 10.1186/s12864-017-4213-5
Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction
Abstract
Background: The plant-specific gibberellic acid stimulated Arabidopsis (GASA) gene family is critical for plant development. However, little is known about these genes, particularly in fruit tree species.
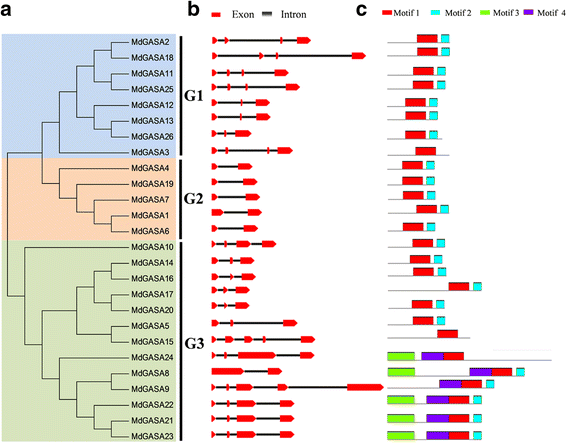
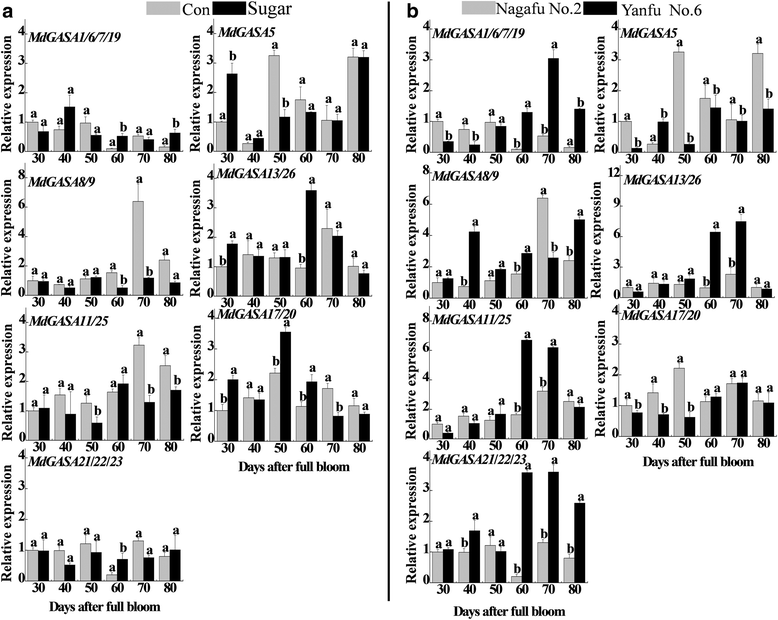
Results: We identified 15 putative Arabidopsis thaliana GASA (AtGASA) and 26 apple GASA (MdGASA) genes. The identified genes were then characterized (e.g., chromosomal location, structure, and evolutionary relationships). All of the identified A. thaliana and apple GASA proteins included a conserved GASA domain and exhibited similar characteristics. Specifically, the MdGASA expression levels in various tissues and organs were analyzed based on an online gene expression profile and by qRT-PCR. These genes were more highly expressed in the leaves, buds, and fruits compared with the seeds, roots, and seedlings. MdGASA genes were also responsive to gibberellic acid (GA3) and abscisic acid treatments. Additionally, transcriptome sequencing results revealed seven potential flowering-related MdGASA genes. We analyzed the expression levels of these genes in response to flowering-related treatments (GA3, 6-benzylaminopurine, and sugar) and in apple varieties that differed in terms of flowering ('Nagafu No. 2' and 'Yanfu No. 6') during the flower induction period. These candidate MdGASA genes exhibited diverse expression patterns. The expression levels of six MdGASA genes were inhibited by GA3, while the expression of one gene was up-regulated. Additionally, there were expression-level differences induced by the 6-benzylaminopurine and sugar treatments during the flower induction stage, as well as in the different flowering varieties.
Conclusion: This study represents the first comprehensive investigation of the A. thaliana and apple GASA gene families. Our data may provide useful clues for future studies and may support the hypotheses regarding the role of GASA proteins during the flower induction stage in fruit tree species.
Keywords: Apple; Characterization; Expression profile; Flower induction; GASA gene.
Conflict of interest statement
Ethics approval and consent to participate
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures









References
-
- Shi LF, Gast RT, Gopalraj M, Olszewski NE. Characterization of a shoot-specific, Ga3-regulated and Aba-regulated gene from tomato. Plant J. 1992;2(2):153–159. - PubMed
-
- Nahirnak V, Rivarola M, de Urreta MG, Paniego N, Hopp HE, Almasia NI, Vazquez-Rovere C. Genome-wide analysis of the Snakin/GASA gene family in Solanum Tuberosum cv. Kennebec. Am J Potato Res. 2016;93(2):172–188. doi: 10.1007/s12230-016-9494-8. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

