Transcriptomic data from two primary cell models stimulating human monocytes suggest inhibition of oxidative phosphorylation and mitochondrial function by N. meningitidis which is partially up-regulated by IL-10
- PMID: 29078758
- PMCID: PMC5659018
- DOI: 10.1186/s12865-017-0229-5
Transcriptomic data from two primary cell models stimulating human monocytes suggest inhibition of oxidative phosphorylation and mitochondrial function by N. meningitidis which is partially up-regulated by IL-10
Abstract
Background: Biological interpretation of DNA microarray data may differ depending on underlying assumptions and statistical tests of bioinformatics tools used. We used Gene Set Enrichment Analysis (GSEA) and Ingenuity Pathway Analysis (IPA) to analyze previously generated DNA microarray data from human monocytes stimulated with N. meningitidis and IL-10 ("the model system"), and with meningococcal sepsis plasma before and after immunodepletion of IL-10 ("the patient plasma system"). The objectives were to compare if the two bioinformatics methods resulted in similar biological interpretation of the datasets, and to identify whether GSEA provided additional insight compared with IPA about the monocyte host response to meningococcal activation.
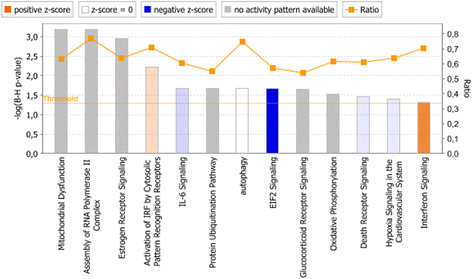
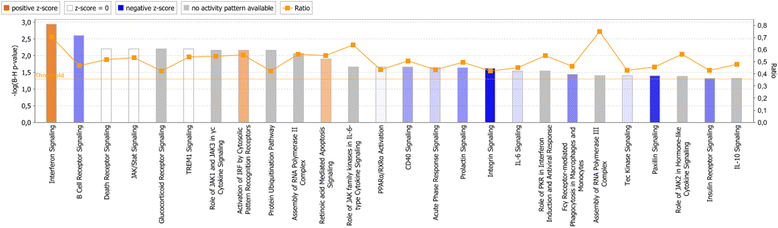
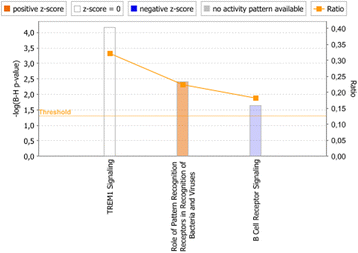
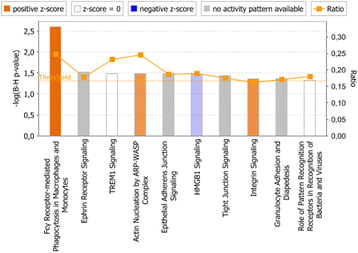
Results: In both experimental models, GSEA and IPA identified genes associated with pro-inflammatory innate immune activation, including TNF-signaling, Toll-like receptor signaling, JAK-STAT-signaling, and type I and type II interferon signaling. GSEA identified genes regulated by the presence of IL-10 with similar gene sets in both the model system and the patient plasma system. In the model system, GSEA and IPA in sum identified 170 genes associated with oxidative phosphorylation/mitochondrial function to be down-regulated in monocytes stimulated with meningococci. In the patient plasma system, GSEA and IPA in sum identified 122 genes associated with oxidative phosphorylation/mitochondrial dysfunction to be down-regulated by meningococcal sepsis plasma depleted for IL-10. Using IPA, we identified IL-10 to up-regulate 18 genes associated with oxidative phosphorylation/mitochondrial function that were down-regulated by N. meningitidis.
Conclusions: Biological processes associated with the gene expression changes in the model system of meningococcal sepsis were comparable with the results found in the patient plasma system. By combining GSEA with IPA, we discovered an inhibitory effect of N. meningitidis on genes associated with mitochondrial function and oxidative phosphorylation, and that IL-10 partially reverses this strong inhibitory effect, thereby identifying, to our knowledge, yet another group of genes where IL-10 regulates the effect of LPS. We suggest that relying on a single bioinformatics tool together with an arbitrarily chosen filtering criteria for data analysis may result in overlooking relevant biological processes and signaling pathways associated with genes differentially expressed between compared experimental conditions.
Keywords: Bioinformatics; Gene expression; Gene set enrichment analysis; Ingenuity pathway analysis; Interleukin-10; Meningococcal sepsis; N. Meningitidis; mRNA.
Conflict of interest statement
Ethics approval and consent to participate
This study used DNA microarray data generated from the use of patient plasma samples (
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

