The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element
- PMID: 29078818
- PMCID: PMC5660458
- DOI: 10.1186/s13059-017-1331-y
The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element
Abstract
Background: It is now obvious that the majority of cellular transcripts do not code for proteins, and a significant subset of them are long non-coding RNAs (lncRNAs). Many lncRNAs show aberrant expression in cancer, and some of them have been linked to cell transformation. However, the underlying mechanisms remain poorly understood and it is unknown how the sequences of lncRNA dictate their function.
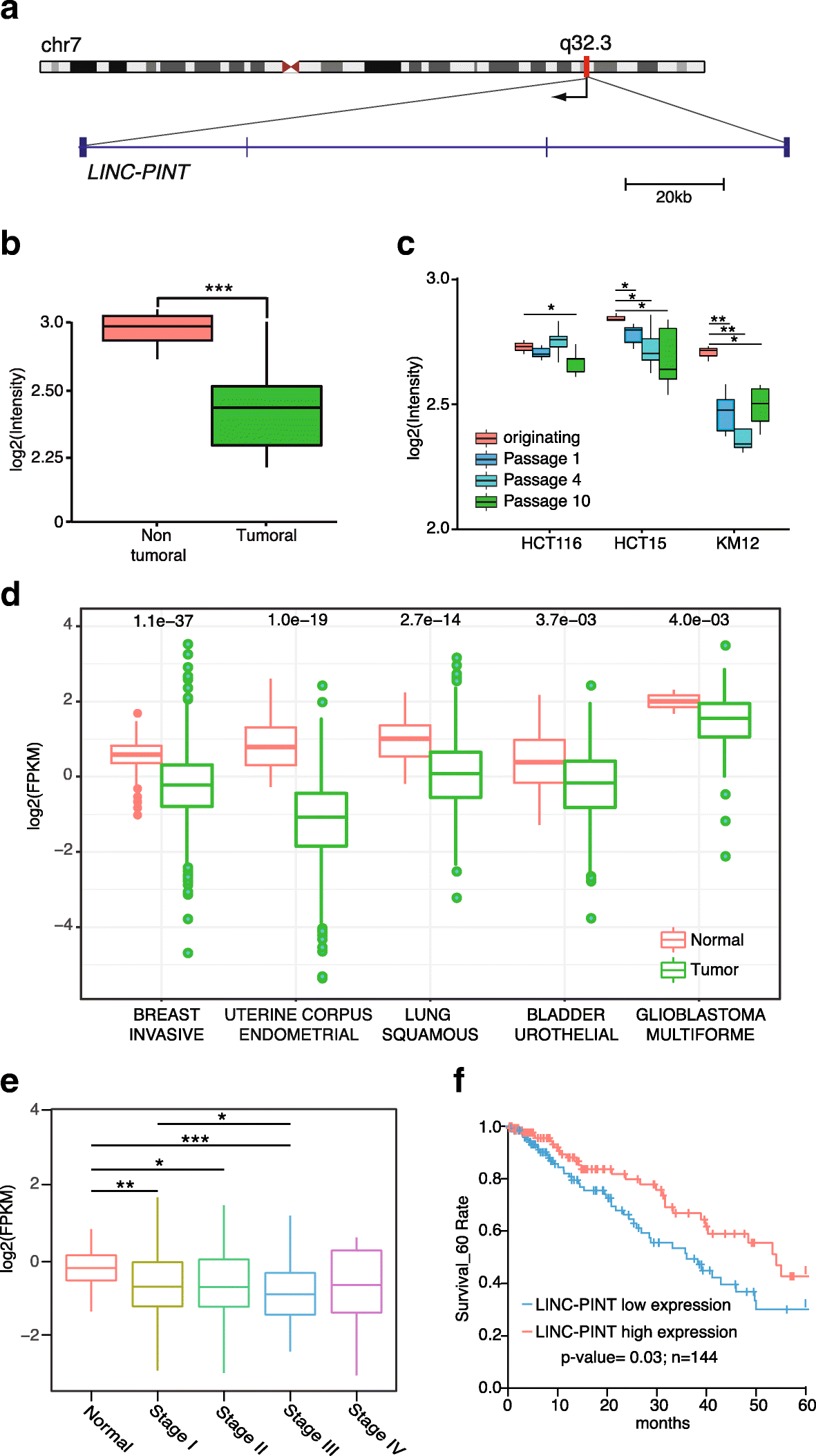
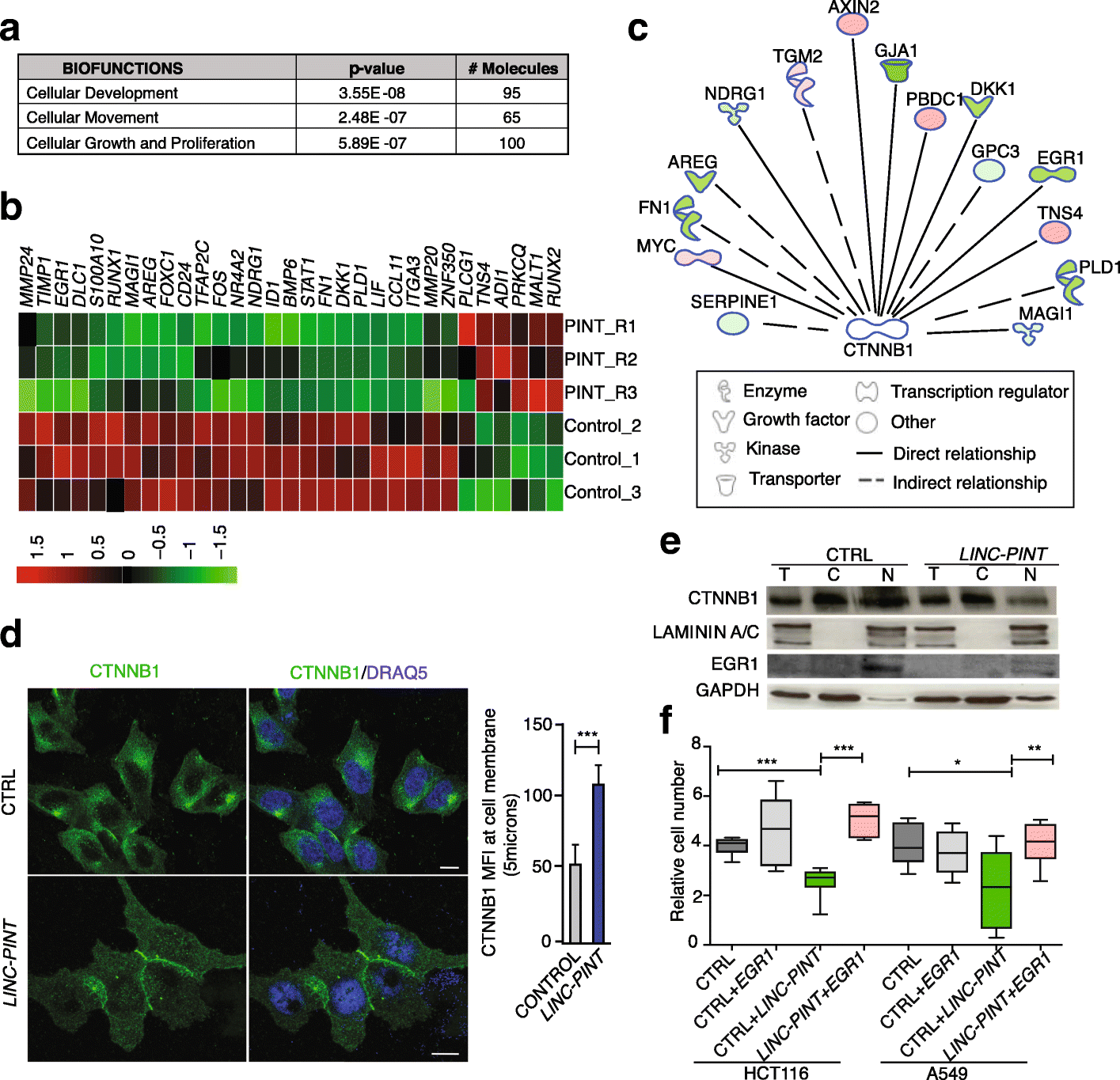
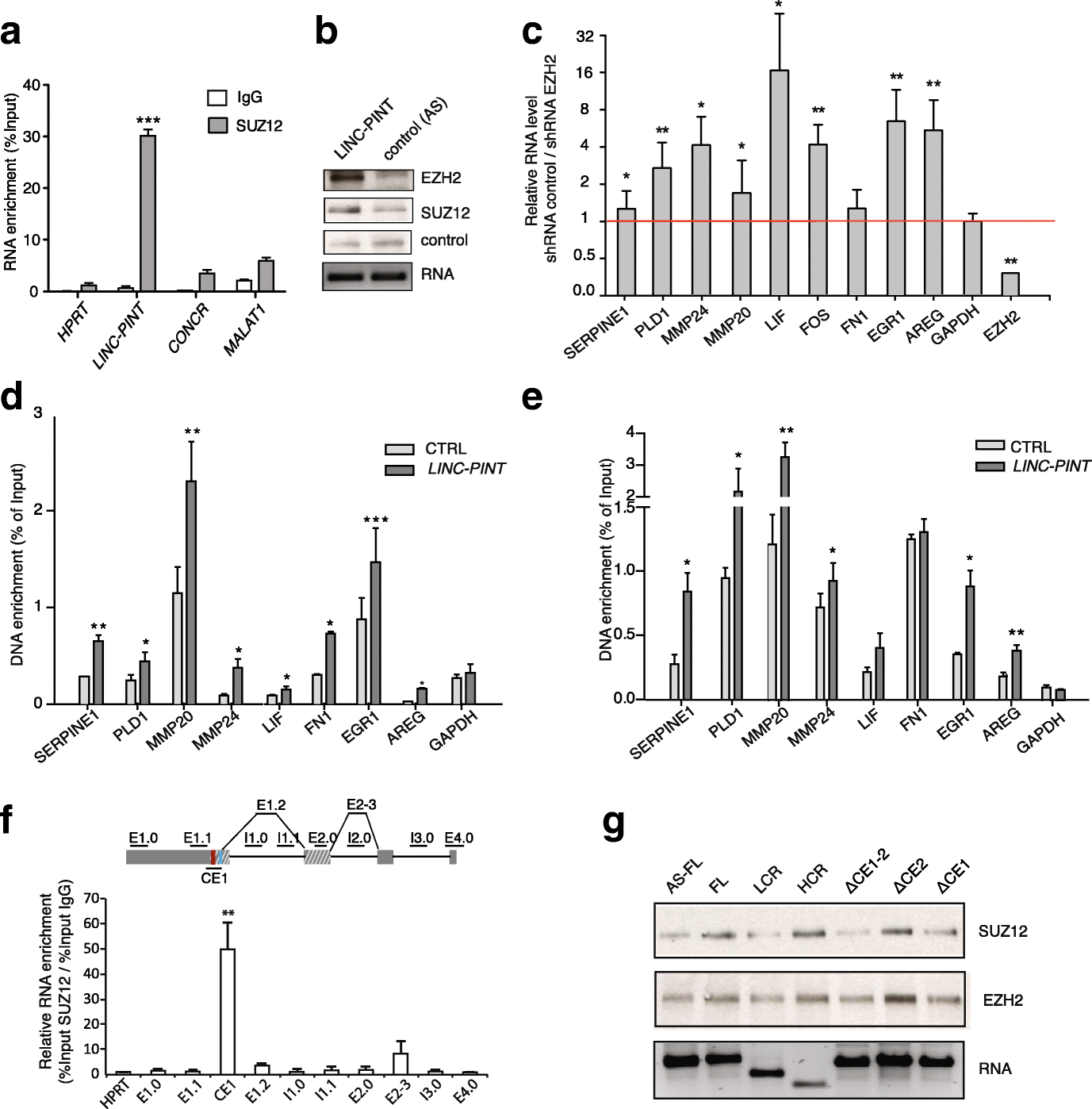
Results: Here we characterize the function of the p53-regulated human lncRNA LINC-PINT in cancer. We find that LINC-PINT is downregulated in multiple types of cancer and acts as a tumor suppressor lncRNA by reducing the invasive phenotype of cancer cells. A cross-species analysis identifies a highly conserved sequence element in LINC-PINT that is essential for its function. This sequence mediates a specific interaction with PRC2, necessary for the LINC-PINT-dependent repression of a pro-invasion signature of genes regulated by the transcription factor EGR1.
Conclusions: Our findings support a conserved functional co-dependence between LINC-PINT and PRC2 and lead us to propose a new mechanism where the lncRNA regulates the availability of free PRC2 at the proximity of co-regulated genomic loci.
Keywords: Cancer; Cell invasion; Epigenetic regulation; LncRNA; PRC2.
Conflict of interest statement
Competing interests
None of the authors have any competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

