Analysis of airborne microbial communities using 16S ribosomal RNA: Potential bias due to air sampling stress
- PMID: 29079080
- PMCID: PMC5805565
- DOI: 10.1016/j.scitotenv.2017.10.154
Analysis of airborne microbial communities using 16S ribosomal RNA: Potential bias due to air sampling stress
Abstract
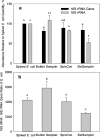
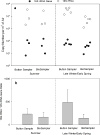
A limited number of studies have been conducted to analyze ribosomal RNA (rRNA, present in the ribosome) in bioaerosol samples to identify currently or potentially active airborne microbes, although its genomic counterpart, the rRNA gene (on the chromosome) has been frequently targeted for airborne microbial community analysis. A knowledge gap still exists regarding whether the bioaerosol rRNA abundances are affected by the bioaerosol collection process. We investigated the effect of air sampling stress on the measurement and characterization of 16S rRNA for bioaerosols in the laboratory and field experiments using quantitative polymerase chain reaction (qPCR) and high-throughput sequencing techniques. In a laboratory study, known quantities of freshly grown Escherichia coli cells were spiked onto the filter of a Button Aerosol Sampler and into liquids of BioSampler and SpinCon air samplers and then exposed to sampling stress when the samplers were operated for 2h. We found that the recovered cellular 16S rRNA abundance as determined by qPCR was dependent on sampler type. Further, two devices (Button Aerosol Sampler and BioSampler) that exhibited markedly different efficiency in preserving 16S rRNA were employed in an outdoor environment to collect bioaerosols simultaneously on eight days in two different seasons. The abundance of 16S rRNA in the outdoor air sample (1.3×106-4.9×107copies/m3) was about two orders of magnitude higher than that of 16S rRNA gene (6.9×103-1.5×105copies/m3). The 16S rRNA sequences revealed a different bacterial community compared with 16S rRNA gene-based results across all samples, and this difference depended on the sampling device. In addition, a number of bacterial taxa exhibited higher abundance in the 16S rRNA gene sequences than in 16S rRNA sequences, which suggests the potential activities of certain microbes in airborne phase. Overall, this study highlights the importance of sampling device selection when analyzing RNA in bioaerosols.
Keywords: 16S rRNA sequencing; Bioaerosols; Microbial community; Sampling device; Stress; rRNA:rDNA ratio.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures



References
-
- Keer JT, Birch L. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods. 2003;53:175–183. - PubMed
-
- England LS, Holmes SB, Trevors JT. Persistence of viruses and DNA in soil. World Journal of Microbiology & Biotechnology. 1998;14(2):163–169.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

