The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity
- PMID: 29079573
- PMCID: PMC5766958
- DOI: 10.1074/jbc.M117.800482
The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity
Abstract
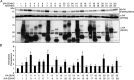
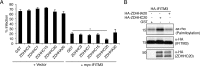
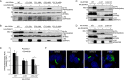
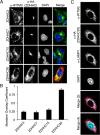
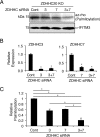
Interferon-induced transmembrane protein 3 (IFITM3) is a cellular endosome- and lysosome-localized protein that restricts numerous virus infections. IFITM3 is activated by palmitoylation, a lipid posttranslational modification. Palmitoylation of proteins is primarily mediated by zinc finger DHHC domain-containing palmitoyltransferases (ZDHHCs), but which members of this enzyme family can modify IFITM3 is not known. Here, we screened a library of human cell lines individually lacking ZDHHCs 1-24 and found that IFITM3 palmitoylation and its inhibition of influenza virus infection remained strong in the absence of any single ZDHHC, suggesting functional redundancy of these enzymes in the IFITM3-mediated antiviral response. In an overexpression screen with 23 mammalian ZDHHCs, we unexpectedly observed that more than half of the ZDHHCs were capable of increasing IFITM3 palmitoylation with ZDHHCs 3, 7, 15, and 20 having the greatest effect. Among these four enzymes, ZDHHC20 uniquely increased IFITM3 antiviral activity when both proteins were overexpressed. ZDHHC20 colocalized extensively with IFITM3 at lysosomes unlike ZDHHCs 3, 7, and 15, which showed a defined perinuclear localization pattern, suggesting that the location at which IFITM3 is palmitoylated may influence its activity. Unlike knock-out of individual ZDHHCs, siRNA-mediated knockdown of both ZDHHC3 and ZDHHC7 in ZDHHC20 knock-out cells decreased endogenous IFITM3 palmitoylation. Overall, our results demonstrate that multiple ZDHHCs can palmitoylate IFITM3 to ensure a robust antiviral response and that ZDHHC20 may serve as a particularly useful tool for understanding and enhancing IFITM3 activity.
Keywords: DHHC; IFITM; IFITM3; ZDHHC; influenza virus; innate immunity; interferon; palmitoyltransferase; post-translational modification (PTM); protein palmitoylation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., and Elledge S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

