Interferon regulatory factor 5 (IRF5) suppresses hepatitis C virus (HCV) replication and HCV-associated hepatocellular carcinoma
- PMID: 29079574
- PMCID: PMC5766933
- DOI: 10.1074/jbc.M117.792721
Interferon regulatory factor 5 (IRF5) suppresses hepatitis C virus (HCV) replication and HCV-associated hepatocellular carcinoma
Abstract
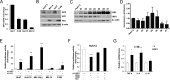
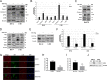
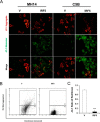
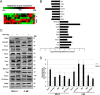
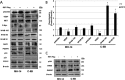
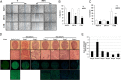
Hepatitis C virus (HCV) infection is a major risk factor for the development of chronic liver disease. The disease typically progresses from chronic HCV to fibrosis, cirrhosis, hepatocellular carcinoma (HCC), and death. Chronic inflammation associated with HCV infection is implicated in cirrhosis and HCC, but the molecular players and signaling pathways contributing to these processes remain largely unknown. Interferon regulatory factor 5 (IRF5) is a molecule of interest in HCV-associated HCC because it has critical roles in virus-, Toll-like receptor (TLR)-, and IFN-induced signaling pathways. IRF5 is also a tumor suppressor, and its expression is dysregulated in several human cancers. Here, we present first evidence that IRF5 expression and signaling are modulated during HCV infection. Using HCV infection of human hepatocytes and cells with autonomously replicating HCV RNA, we found that levels of IRF5 mRNA and protein expression were down-regulated. Of note, reporter assays indicated that IRF5 re-expression inhibited HCV protein translation and RNA replication. Gene expression analysis revealed significant differences in the expression of cancer pathway mediators and autophagy proteins rather than in cytokines between IRF5- and empty vector-transfected HCV replicon cells. IRF5 re-expression induced apoptosis via loss in mitochondrial membrane potential, down-regulated autophagy, and inhibited hepatocyte cell migration/invasion. Analysis of clinical HCC specimens supports a pathologic role for IRF5 in HCV-induced HCC, as IRF5 expression was down-regulated in livers from HCV-positive versus HCV-negative HCC patients or healthy donor livers. These results identify IRF5 as an important suppressor of HCV replication and HCC pathogenesis.
Keywords: Hepatitis C virus (HCV); hepatocellular carcinoma; immunosuppression; interferon regulatory factor (IRF); tumor suppressor gene; viral immunology.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Hajarizadeh B., Grebely J., Dore G. J. (2013) Epidemiology and natural history of HCV infection. Nat. Rev. Gastroenterol. Hepatol. 10, 553–562 - PubMed
-
- Mohd Hanafiah K., Groeger J., Flaxman A. D., and Wiersma S. T. (2013) Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57, 1333–1342 - PubMed
-
- Arzumanyan A., Reis H. M., and Feitelson M. A. (2013) Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Cancer 13, 123–135 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

