Transition of colistin dependence into colistin resistance in Acinetobacter baumannii
- PMID: 29079752
- PMCID: PMC5660220
- DOI: 10.1038/s41598-017-14609-0
Transition of colistin dependence into colistin resistance in Acinetobacter baumannii
Abstract
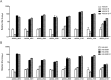
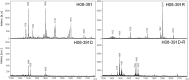
We recently demonstrated a high rate of colistin dependence in Acinetobacter baumannii isolates exposed to colistin in vitro. In the present study, we obtained a colistin-resistant (H08-391R) and colistin-dependent mutant (H08-391D) from a colistin-susceptible parental strain (H08-391). We found that the colistin-dependent mutant converted into a stable colistin-resistant mutant (H08-391D-R) in vitro after four serial passages without colistin. H08-391D and H08-391D-R were both found to harbor defective lipid A, as indicated by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry analysis. Additionally, both contained an ISAba1 insertion in lpxC, which encodes a lipid A biosynthetic enzyme. Further, membrane potential measurements using the fluorescent dye 3,3'-diethyloxacarbocyanine iodide (DiOC2[3]) showed that the membrane potential of H08-391D and H08-391D-R was significantly decreased as compared to that of the parental strain, H08-391. Moreover, these mutant strains exhibited increased susceptibilities to antibiotics other than colistin, which may be attributed to their outer membrane fragility. Such phenomena were identified in other A. baumannii strains (H06-855 and its derivatives). Taken together, our study reveals that the colistin-dependent phenotype is a transient phenotype that allows A. baumannii to survive under colistin pressure, and can transition to the extremely resistant phenotype, even in an antibiotic-free environment.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

