Comparative analysis of mitochondrial genomes of geographic variants of the gypsy moth, Lymantria dispar, reveals a previously undescribed genotypic entity
- PMID: 29079798
- PMCID: PMC5660218
- DOI: 10.1038/s41598-017-14530-6
Comparative analysis of mitochondrial genomes of geographic variants of the gypsy moth, Lymantria dispar, reveals a previously undescribed genotypic entity
Abstract
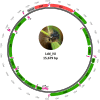
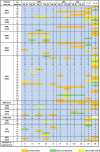
The gypsy moth, Lymantria dispar L., is one of the most destructive forest pests in the world. While the subspecies established in North America is the European gypsy moth (L. dispar dispar), whose females are flightless, the two Asian subspecies, L. dispar asiatica and L. dispar japonica, have flight-capable females, enhancing their invasiveness and warranting precautionary measures to prevent their permanent establishment in North America. Various molecular tools have been developed to help distinguish European from Asian subspecies, several of which are based on the mitochondrial barcode region. In an effort to identify additional informative markers, we undertook the sequencing and analysis of the mitogenomes of 10 geographic variants of L. dispar, including two or more variants of each subspecies, plus the closely related L. umbrosa as outgroup. Several regions of the gypsy moth mitogenomes displayed nucleotide substitutions with potential usefulness for the identification of subspecies and/or geographic origins. Interestingly, the mitogenome of one geographic variant displayed significant divergence relative to the remaining variants, raising questions about its taxonomic status. Phylogenetic analyses placed this population from northern Iran as basal to the L. dispar clades. The present findings will help improve diagnostic tests aimed at limiting risks of AGM invasions.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Pogue, M. & Schaefer, P. W. A review of selected species of Lymantria Hübner (1819) (Lepidoptera: Noctuidae: Lymantriinae) from subtropical and temperate regions ofAsia, including the descriptions of three new species, some potentially invasive to North America (U.S. Dept. of Agriculture, Forest Health Technology Enterprise Team, Washington, D.C. 2007).
-
- Forbush, E. H. & Fernald, C. H. The gypsy moth, Porthetria dispar (Linn.): a report of the work of destroying the insect in the Commonwealth of Massachusetts, together with an account of its history and habits both in Massachusetts and Europe. (Wright & Potter Printing Co., Boston, 1896).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

