Comparative analysis of murine T-cell receptor repertoires
- PMID: 29080364
- PMCID: PMC5765371
- DOI: 10.1111/imm.12857
Comparative analysis of murine T-cell receptor repertoires
Abstract
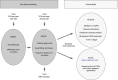
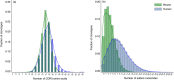
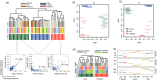
For understanding the rules and laws of adaptive immunity, high-throughput profiling of T-cell receptor (TCR) repertoires becomes a powerful tool. The structure of TCR repertoires is instructive even before the antigen specificity of each particular receptor becomes available. It embodies information about the thymic and peripheral selection of T cells; the readiness of an adaptive immunity to withstand new challenges; the character, magnitude and memory of immune responses; and the aetiological and functional proximity of T-cell subsets. Here, we describe our current analytical approaches for the comparative analysis of murine TCR repertoires, and show several examples of how these approaches can be applied for particular experimental settings. We analyse the efficiency of different metrics used for estimation of repertoire diversity, repertoire overlap, V-gene and J-gene segments usage similarity, and amino acid composition of CDR3. We discuss basic differences of these metrics and their advantages and limitations in different experimental models, and we provide guidelines for choosing an efficient way to lead a comparative analysis of TCR repertoires. Applied to the various known and newly developed mouse models, such analysis should allow us to disentangle multiple sophisticated puzzles in adaptive immunity.
Keywords: T cell; T-cell receptor repertoires; aging; diversity; functional T-cell subsets.
© 2017 John Wiley & Sons Ltd.
Figures




References
-
- Linnemann C, Mezzadra R, Schumacher TN. TCR repertoires of intratumoral T‐cell subsets. Immunol Rev 2014; 257:72–82. - PubMed
-
- Greiff V, Miho E, Menzel U, Reddy ST. Bioinformatic and statistical analysis of adaptive immune repertoires. Trends Immunol 2015; 36:738–49. - PubMed
-
- Heather JM, Ismail M, Oakes T, Chain B. High‐throughput sequencing of the T‐cell receptor repertoire: pitfalls and opportunities. Brief Bioinform 2017; doi: 10.1093/bib/bbw138. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

